|
| |
|
Main Newsletter Index
AIPS++ for
|
Msplot Tim Cornwell (revised by Ralph Marson) - NRAO/Socorro The msplot tool is currently the main interactive tool for displaying and editing data from an AIPS++ MeasurementSet. It is written as an AIPS++ tool using a number of lower-level tools such as ms, pgplotter, and the viewer. An msplot tool can be constructed either using the toolmanager or from the command line using:
mymsplot := msplot('mydata.ms')
However if you wish to flag data in the measurement set you should use:
mymsplot := msplot('mydata.ms', edit=T)
Either of these commands will start a graphical interface to mymsplot allowing the user to display and optionally edit the data in the measurement set. Currently, msplot is entirely GUI-based so that none of the operations can be invoked from the command line.
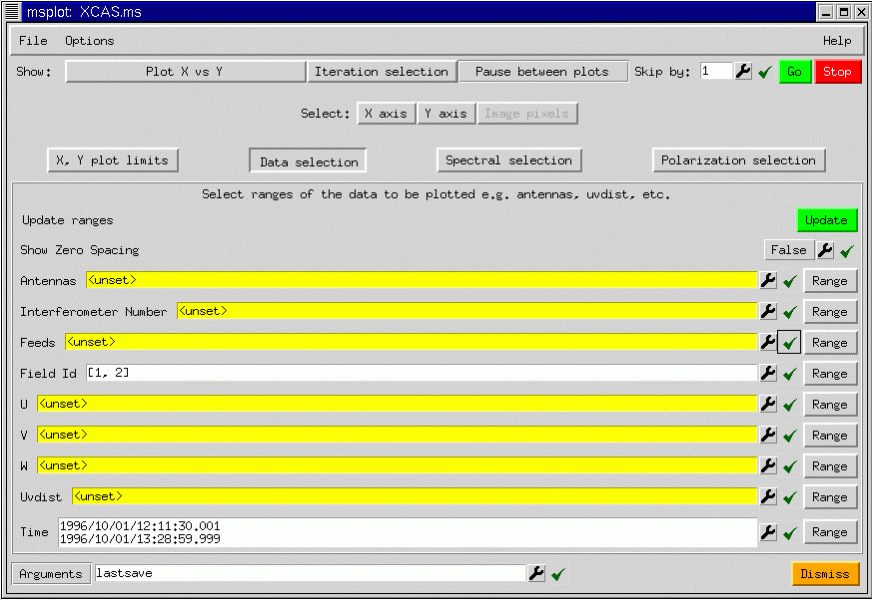
The GUI shows a number of buttons controlling the data selection and the type of plot or display produced. What data is plotted and how it is displayed can be chosen as follows:
The axes may be chosen as follows:
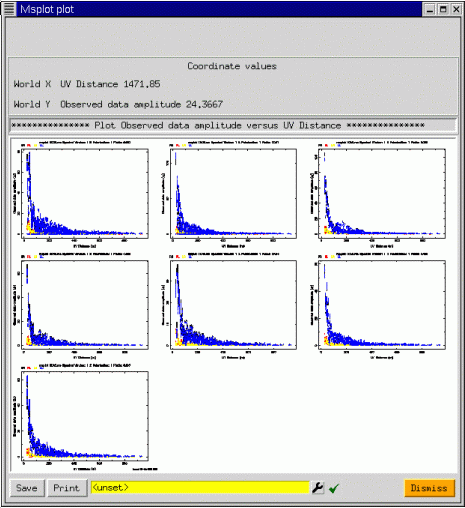
In addition to these axis selections, msplot can iterate over various indices such as antenna, feed, field, scan number, spectral window, and polarization. As you can see, the possibilities are vast! To show a simple plot, taking advantage of the iteration capabilities, we show msplot applied to a seven pointing mosaic data set from the VLA. We plot all corrected visibility amplitudes versus uv distance iterating over field:
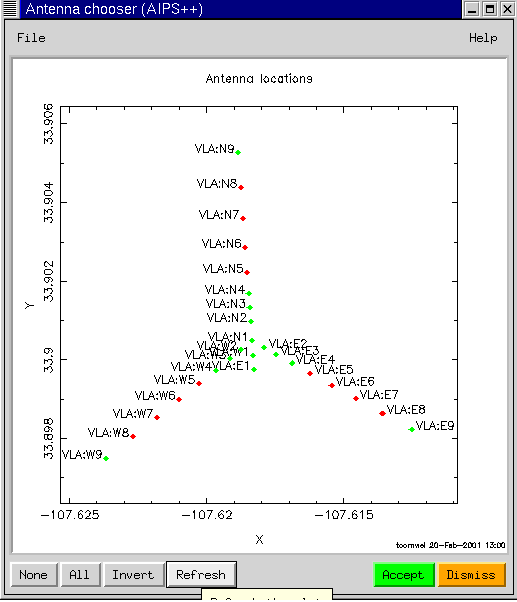
Suppose that we wish to see just the visibilities from the inner antennas for field 1. Further selection of the data can be performed using the Data selection button. We press the Data selection button to display the GUI as shown above. We can either type in the Antenna numbers, or better, select them graphically by pressing on the spanner (or wrench) for the Antennas argument.
One can select the antennas to be plotted by dragging a box. The corresponding numbers in the Antennas argument are then filled in when the Accept button is pressed. Similar graphical elements (gophers in AIPS++) can be used to get other types of information. If we narrow down the fields to show only the first field, and repeat the plot, we get:
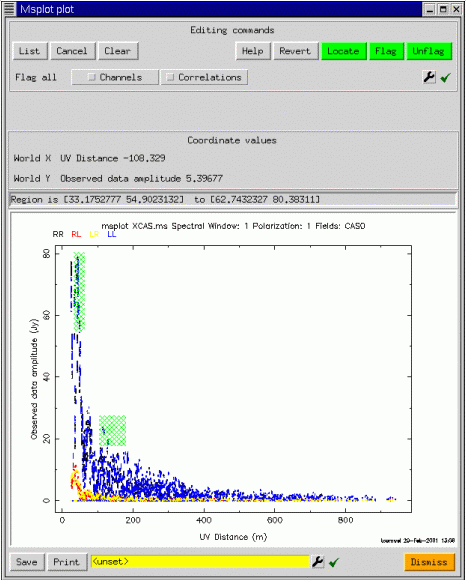
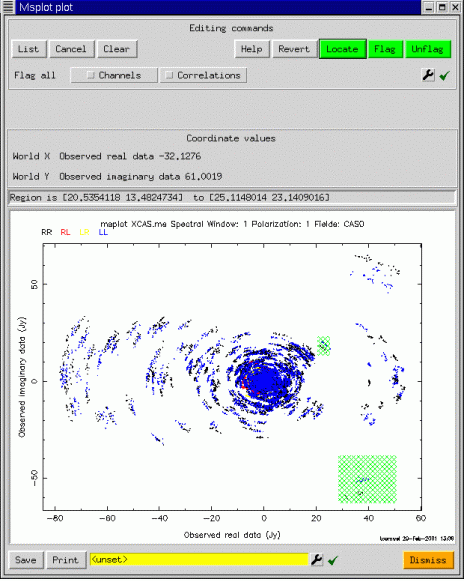
Now since we have only one plot, editing is enabled (assuming msplot was started with edit=T). To edit points, one uses a click and drag action to select the points (shown in hatched green), followed by a press of the Flag button (top right), to actually edit the points. The plot is then repeated, and the corresponding points are erased. Editing is done in place on the FLAG and FLAG_ROW columns of the measurement set. At startup, these current columns are saved to a scratch table, from whence they may be restored if the user decides not to apply the edits. One of the unexpectedly powerful features of msplot is that editing is possible in any scatter plot. For example, for the same selection, we may edit in the plot of real versus imaginary part of the visibility:
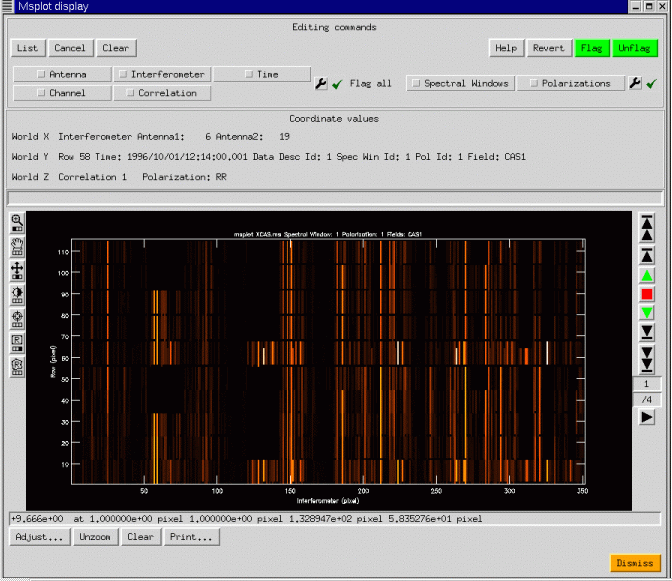
Although we have concentrated on scatter plots, image-like displays are also possible. msplot uses the viewer to provide the image display capabilities, and so the interface shown below should have some familiar aspects:
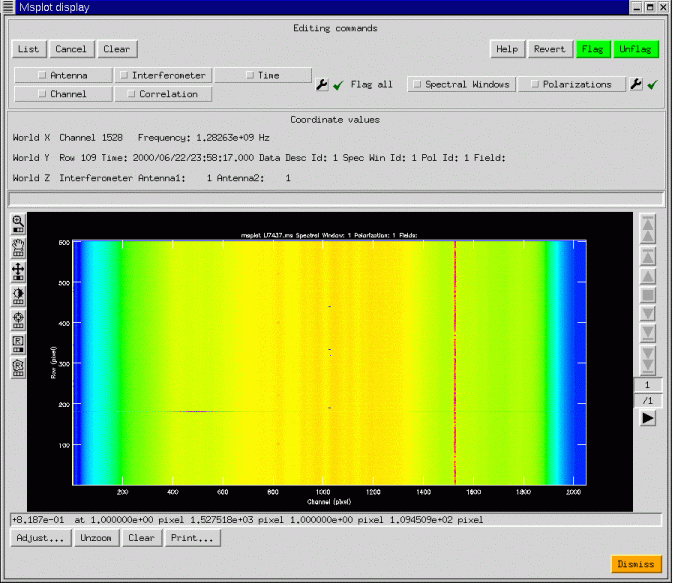
We have shown a simple continuum display, with 4 correlations (RR, RL, LR, LL). The ability of AIPS++ to access with equal efficiency along any set of image axes can be used to good effect: the data are gridded into an image with axes Interferometer, Time, Channel, and Polarization. Any two of these may be chosen as the first two axes on the display, and a third scrolled through in a movie. In the example above, the four correlations are shown as the third axis through which the viewer may be scrolled. Editing is possible in the display mode and is preferable to the plotting mode when the data volume is large. Msplot can also display single dish data with the same wide range of possibilities. In the next figure, we show an image display of some 20cm data from the Arecibo telescope complete with narrow-band interference.
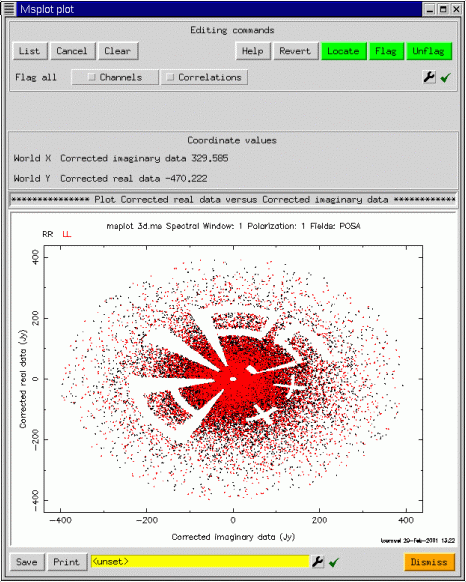
This is only a brief tour of the capabilities of the msplot tool. It seems to be popular for data editing, once people get accustomed to the flexibility of the interface. I recommend that you experiment quite a bit since some of the more esoteric types of plot can provide excellent diagnostic capabilities. msplot was designed and developed as a prototype, and it is expected that many of the capabilities now provided by the pgplotter and the viewer will instead be offered as native capabilities of the viewer as the continuing development of the Display Library allows. Finally, we end with another plot on a 327MHz VLA measurement set, showing how the various capabilities described here can be combined. We'll let the reader figure out how this was done.
|
|
Main Newsletter Index
AIPS++ for
|
Image Mask Handling Neil Killeen - ATNF
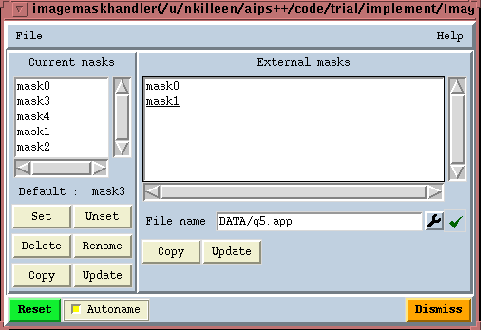
GeneralEach pixel in an image has a brightness value and a mask value. A good pixel (mask value True) is one that the image analysis software will include in its computations or displays. A bad pixel (mask value False) is one that will be excluded from calculations. We store these Boolean mask values in what is termed the 'pixel mask'. An image may hold zero, one or more pixel masks. This is rather handy as it allows you to try many things out without having to continually overwrite the mask. These masks are identified by name. The Image tool function summary lists them for you. Here is an example.
Image name : zz Image type : PagedImage Pixel mask(s) : mask2 [mask0, mask1] Region(s) : None Image units : Jy/beam Restoring Beam : 53.5 arcsec, 34.2 arcsec, 6 deg Direction reference : J2000 Axis Type Name Proj Shape Tile Coord value at pixel Coord incr Units -------------------------------------------------------------------------------- ---------- 1 Direction Right Ascension SIN 113 113 00:00:00.000 56.00 -8.000000 e+00 arcsec 2 Direction Declination SIN 76 76 +00.00.00.000 38.00 1.200000 e+01 arcsec You can see that this image has three masks. The first one, 'mask2' is currently the active mask. This means it will be applied by default by all the analysis software. The other two, listed in square brackets, are not currently active. If none of the masks were active, they would all be listed in square brackets. This means that effectively an 'all good' mask is applied.
Managing the pixel masks in an imageThere is an Image tool function called maskhandler. Its job is to let you manipulate masks (e.g. copy, delete etc.). Its job is not to change the pixel mask values (see following subsection). Its interface is
- im := image('myimage')
- im.maskhandler(op, name)
where op is a string specifying an operation, and name is a string argument that may be required depending on the chosen operation. The argument op may take the values (minimum match allowed) :
As well as the command-line interface, there is also a custom GUI for the maskhandler function. It is available from the Toolmanager as well as as via an Image tool command-line function.
g := im.maskhandlergui() # g is a tool itself
The left-hand list box shows you the pixel masks currently available in the image. Underneath the list box the default pixel mask, if any, is listed. You have buttons under the left-hand list box to apply the desired operation. The sequence is to select a pixel mask or pixel masks from the list box, and then click the desired button. Popuphelp is on each button to tell you what they do. The right-hand list box is used to list the pixel masks from some other image. You enter the image file name with the file entry widget and then enter carriage-return. You can copy masks from that image file to the current image.
Changing the values of pixel masksThere are a few ways in which you can change the values of a pixel mask.
The mask argumentThere is an argument, mask, which can be supplied to many Image tool constructors and functions. It is supplied with a LEL Boolean expression string. One example of its use is in function calcmask described above where the resultant mask is stored with the image. In all other uses, the mask argument is used to generate an On-The-Fly (OTF) mask. It is generated, used, and discarded once the function terminates. In the case of calcmask, its use is to store it, rather than apply it for some other analysis. The OTF mask is applied in addition to any default mask the image holds. Here are some examples:
- im := imagemaketestimage('zz')
- im.statistics(mask='$im > 0') # Stats evaluated when default mask of $im a
nd OTF mask are True
- im.view (mask='yy<0') # Displayed when default mask of $im and OTF
mask are True
- im.moments(mask='abs(zz)>3*stddev(zz)')
It is especially handy in the view function.
Replacing masked pixelsEach pixel has a brightness value and a mask value. Sometimes you may want to replace the value of all masked pixels, and then delete the mask. This is done with the Image tool function replacemaskedpixels. The argument pixels can take a scalar, a scalar LEL expression or a non-scalar LEL expression of the correct shape.
- im := imagemaketestimage('zz')
- im.calcmask('zz>0')
- im.replacemaskedpixels(pixels='-1*$im') # All masked pixels replaced by the
ir negative
- im.replacemaskedpixels(pixels=0) # All masked pixels replaced by 0
- im.replacemaskedpixels(pixels='min(yy)') # All masked pixels replaced by the
minimum of image yy
- im.maskhandler('set','') # Unset default mask
You can also use the OTF mask argument if you wish. Note that in this example no actual persistent mask is created.
- im := imagemaketestimage('zz') # Has no mask
- im.replacemaskedpixels(pixels=0, mask='zz>0') # All non-positive values s
et to 0
Pixel masks and regionsSome comment about the combination of pixel masks and regions-of-interest is useful here. See the Regionmanager tool for basic information about regions-of-interest first. Regions are provided to Image tool functions via the standard region function argument. Consider a simple polygonal region. This region-of-interest is defined by a bounding box, the polygonal vertices, and a mask called a 'region mask'. The region mask specifies whether a pixel within the bounding box is inside or outside the polygon. For a simple box region-of-interest, there is obviously no need for a region mask. Now imagine that you wish to recover the pixel mask of an image from a polygonal region-of-interest. The mask is returned to you in regular Boolean array. Thus, the shape of the returned mask array reflects the bounding-box of the polygonal region. If the actual pixel mask that you apply is all good, then the retrieved mask would be good inside of the polygonal region and bad outside of it. If the actual pixel mask had some bad values in it as well, the retrieved mask would be bad outside of the polygonal region. Inside the polygonal region it would be bad if the pixel mask was bad. More simply put, the mask that you recover is just a logical ``and'' of the pixel mask and the region mask; if the pixel mask is True and the region mask is True then the retrieved mask is True (good), else it is False (bad). Finally, note that if you use the region and mask (the OTF mask) arguments together then they operate as follows. The shape of the Boolean expression provided by mask must be the same shape as the image to which it is being applied. The region is applied equally to the image and the mask expression. For example
- rm1 := image('rm')
- rm2 := image('rmerr')
- rm1.shape(); rm2.shape()
[128 128]
[128 128]
- r := drm.box([10,10], [50,50])
- rm1.statistics(region=r, mask='rmerr<10') # region applied to 'rmerr'
and 'rm'
|
|
Main Newsletter Index
AIPS++ for
|
AIPS++ Viewer Update Malte Marquarding - ATNF What's NewThe aips++ viewer tool is expanding it's functionality. Since the last release following new features have been added or enhanced:
Worked Examples
SkycatalogA Skycatalog is an aips++ table in a specialized format This section describes how to use Skycatalog to plot itself on data of different Direction Coordinate types (J2000, B1950, etc.) simultaneously. This is useful if for example one wants to see where a source is on an HI map and at the same time on the sky in galactic coordinates. The Skycatalog input source might come from a catalog retrieved from a database and the source coordinates can be of any direction type. There is tools to simplify creating these tables, the skycatalog tool.
Using Skycatalog to annotate imagesThe Skycatalog can be used as well to annotate an image.We start with creating an (empty) skycatalog tool.
include 'skycatalog.g';
sc := skycatalog('mycatalog.tbl');
Now we make an image tool, in this case we use the built-in test
image.
include 'image.g'
im := imagemaketestimage('myimage.im');
Now we use Image.findsources() to get
the positions of our sources:
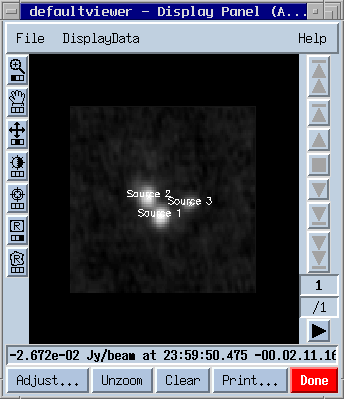
cl := im.findsources();Now we can fill our skycatalog table with the detected components: sc.fromcomponentlist(cl);We have set up a Skycatalog table! It consists of the following columns:
sc.done(); #done with this tool, the table is on disk mycatlog.tbl
dp := dv.newdisplaypanel(); # create a Display Panel
dd := dv.loaddata(im,'raster'); # load our image
dt := dv.loaddata('mycatalog.tbl','skycatalog'); # load our table
# register the image and table
dp.register(dd);
dp.register(dt);
# make the Annotation string visible and make the font bigger
# This can also be done in the Adjust gui
dt.setoptions([namecolumn=[value="Annotation"],labelcharsize=[value=1.5]]);
The result looks something like
Now the overlay can be fine tuned using the Adjust... panel, e.g. different colors, offset from the original position etc. It's best to play around with the available options. If the selected names Src1/2/3 sound too boring use tbl.browse() to edit the fields in the annotation column. To be able to change the table you have to delete the old Sky catalog overlay first: dt.done(); # remove itWe now use the table tool to modify the skycatalog table.
tbl := table('mycatalog.tbl',readonly=F);
tbl.putcell('Annotation',1,'Source A');
tbl.putcell('Annotation',2,'Source B');
tbl.putcell('Annotation',3,'Source C');
tbl.flush();
And after changing the table fields:
dt := dv.loaddata(tbl,'skycatalog'); # load again #set the old options dt.setoptions([namecolumn="Annotation",labelcharsize=1.5]); dp.register(dt);
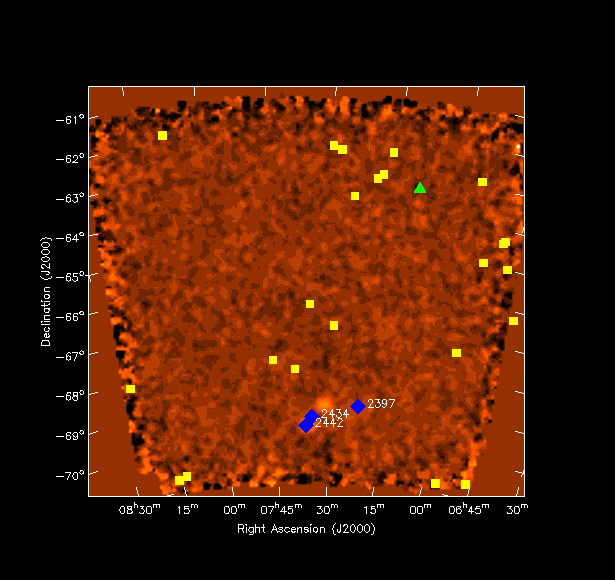
ESOLV catalog converted to skycatalogHere is an example how skycatalog is used to plot source positions from the ESO-LV catalog . The skycatalog in this case was created using the Skycatalog.fromascii function. I have created three catalogs for different arbitrary redshift ranges - column ``CZ'' (green triangles, yellow boxes and blue diamonds)using the Table.query function. For the blue diamonds the table column ``NGC'' was also enabled showing NGC2442, NGC2434 and NGC2397.
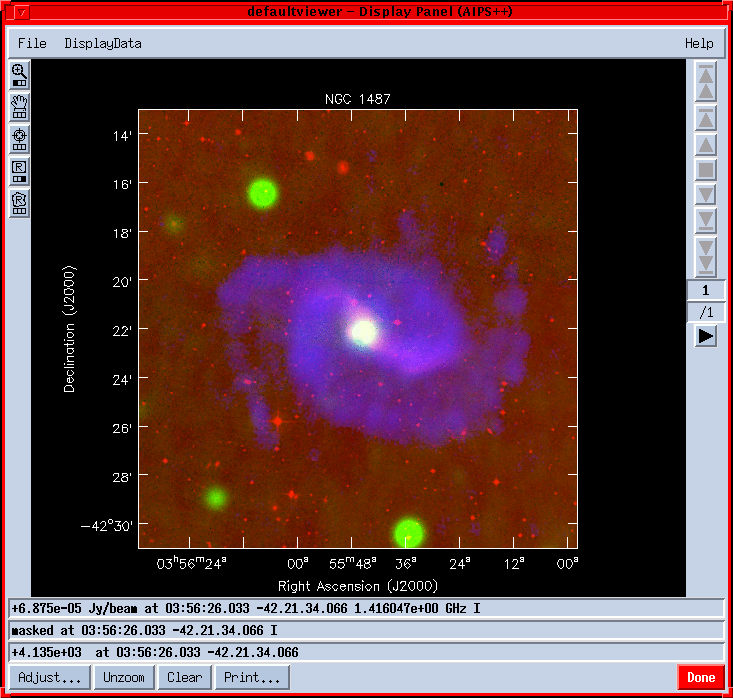
Making RGB multi-frequency overlaysMulti-frequency observations can be easily viewed using the viewer. After starting the viewer with dv.gui(), Done the default Display Panel. Select ``New Display: RGB mode'' from the File menu on the Data Manager. Select your data in the Data Manager and make Raster Images. Even after Registering the DisplayDatas on the Display Panel they won't be visible, because the default is to display themselves in Colormap mode not in R(ed),G(reen) or B(lue). Hit the Adjust button and select one of red, green or blue under ``Basic settings - Color Mode'' for each image. The result can look like :
|
|
Main Newsletter Index
AIPS++ for
|
Parallelization of Imager for the Wide-Field Case K. Golap, A. Kemball, T. Cornwell, W.Young - NRAO/Socorro Wide-field imaging is among the most computationally demanding problems in Radio Astronomy. The wait for Moore's law to help is overtaken by the need for better images, that is more sensitivity and higher resolution. The wide-field imaging problem is due to a geometric effect. Image synthesis arrays can appear to have different geometrical shape from different points in the sky. It is an issue only when this happen within the field of view of the array. This happens in non co-planar arrays and at relatively low frequencies because the primary beam is large. Visibilities as observed by non co-planar is given by
where I(l,m) represents the sky brightness (or image) we want to recover. There are several algorithms that exists to invert this 3-D integral (Cornwell & Perley, 1992). In AIPS++ a multi-faceted transform algorithm has been chosen for its efficiency. This covers the region to be imaged by a series of facets, in each of which a 2-D inversion can be achieved. We can decompose the visibilities into a summation of re-phased faceted visibilities:
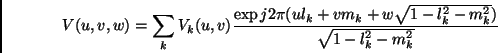
where :
The algorithm implemented in AIPS++ proceeds as follows
What has been parallelized so far in wide-field imaging: We have targeted, as a first level of parallelization, the sections which are nearly "embarassingly parallel". These are the PSF estimations, the model visibility calculation from the model images and the residual image for each facet. We are using a message passing model of a master controller CPU and worker processors as implemented in MPI version 1 libraries. This is implemented on both clusters of workstations and on shared memory multi-processors architectures. The handling of the parallel imager (pimager) is the same as imager and it will run on a single CPU or multi-CPU machines. The only input difference that it needs as one of its inputs the number of free CPU's it has available for its functioning.
Some results of speed ups on NCSA's SGI 2000. The data set used has
1 CPU Future work:
This work is done in collaboration with NCSA's Alliance.
References: Cornwell, T. J., and Perley, R. A., Astron. & Astrophys, 261, 353-364, 1992. Sault, R. Staveley-Smith, L., and Brouw, W. N., Astron. & Astrophys. Suppl., 120, 375-384, 1996.
|
|
Main Newsletter Index
AIPS++ for
|
The VLA Filler Ralph Marson - NRAO/Socorro Reading VLA archive format data into AIPS++The vlafiller is a tool that is used to convert data in VLA archive format into an measurement set. VLA archive format is the native data format that is produced by the online computers at the VLA. All VLA observations are archived, on tape, in Socorro using this data format (hence its name). The measurement set is the data format used by aips++ to store, on disk, the data observed by a telescope. An overview of this data format is in the previous newsletter (November 2000). The vlafiller tool tool performs a similar role to the AIPS task FILLM and has been available to users since the first public release of aips++ (version 1.2). Since then it has undergone a number of significant improvements and is now the preferred method for converting VLA data into a form that can be used within aips++. The alternative is to use the AIPS to convert the VLA archive data into a UV-FITS file that can then be read into AIPS++ using the ms tool. The vlafiller tool can be used in two ways. The simpler way allows you to do the most common tasks ie., reading a tape and creating a measurement set, without any of the bother of having to create a vlafiller tool and remembering to destroy it when you are done. For example if you have a tape with data in VLA archive format you would type at the glish prompt.
- include 'vlafiller.g' #1
- vlafillerfromtape('vla.ms', '/dev/nst0') #2
The first of these lines makes all the vlafiller functions available and the second copies all the data from the tape device to the measurement set. Sometimes you may have a tape with data from other projects besides your own. To only copy the data from your project (AR277) you use the optional project argument and replace the second line above with.
- vlafillerfromtape('ar277.ms', '/dev/nst0', project='AR277')
otherwise you will copy the data from all projects into the measurement set. The vlafiller can also 'filter' the data on a number of other parameters besides the project name. These include time-range, frequency-range, source-name, sub-array, and calibrator-code. The vlafiller can also read VLA archive data from disk, from multiple files on a tape and from tapes on a remote machine. If you create the vlafiller tool yourself (as distinct from using the vlafillerfromtape or vlafillerfromdisk functions) you can do more complex filtering of the data. All these features and more are discussed in the AIPS++ users reference manual in the nrao.vla.vlafiller section.
|
|
Main Newsletter Index
AIPS++ for
|
Using AIPS++ for Amateur Radio Astronomy Patrick Wood - Lobert Observatory, Hinesville, Ga I run an amateur radio observatory from my home. I named my setup LoBERT, Low Budget Experimental Radio Telescope, because so far the total price of my system, including the data logging computer, is under $300. I started the system with just one 15 foot (4.5 meter) dish, in drift scan mode and an old DSS LNBF. I can only control the elevation of the dish, so I have to set the dish to the elevation that I would like to observe that evening and allow the Earth's rotation to act as my RA drive. The telescope is aligned as close to due south as I can get it. I have run multiple scans of the sun and found that the dish is approximately 2 minutes West of due south. I plan on fixing this as soon as I can. I bought an 8 GHz satellite signal meter to use as a total power receiver. I run the meter's DC out for the built in buzzer to a Radio Shack DMM, and wrote a couple of programs to do the data logging. I tried for quite a while to develop my own data reduction software, but to no avail. I found the NRAO web site and found out that they were giving away copies of the AIPS++ system. For me this was like a dream come true, to be able to run the same software as "The Big Boys". I will try and explain how I got my little system to interface with the AIPS++ system. The original configuration of my system was as described above. I use an Intel 486DX4-100 running Windows 95 to do data logging. I can adjust the integration time from 1 second and up. I currently only go to a maximum of 6 seconds. With the current setup the half beam width of LoBERT is approximately .5 degrees. Now you have to remember that all of these calculations are coming from a Food Operations Sergeant in the US Army. I have the data logger and my Linux box on a LAN. I also have another old 486DX-25 running Windows 3.1 running as my sky chart system. I use it to try and figure out exactly what the scope was looking at. Since I have three different OS running together to obtain a common goal, I had figure out a way to get the systems to talk to each other. The way I choose to do this by using the built in Samba networking in Linux. This simplified the connections to the two Windows machines. The data logging computer is running a program developed by a guy named Ned Lewis. His original plan for the software was to do HEPS, High Energy Pulse Surveys, with as many amateur radio astronomers as possible. I got a copy of the program, which is written in Qbasic, and made a few modifications to it. It allows me to run two dishes at the same time and record the data from both dishes at the same sampling rate. The program stores it's data in a MS-DOS ASCII file that looks like the following:
12000 1.329 1.067 12001 1.328 1.069 12002 1.328 1.069 Column1 is the number of seconds that have pased since midnight LST, column2 is the reading in DC volts from the 8' dish, and column3 is the reading from the 15 footer in DC volts. Everyday at midnight LST, the data logger transfers the collected data to the Linux box via the LAN. As I am sure you all know, MS-DOS ASCII and Unix ASCII are two different animals. To solve this program I got hold of the Linux based dos2unix program which converts the ASCII file into something the Linux box can understand. Since I haven't written a program for the Linux box yet, to tell LST I added the following to the crontab:
0 1 * * * dos2unix /home/pw/*.LBR This allows the Linux box to automatically translate the DOS file to an Unix file at 1 o'clock in the morning every morning. Everything up to this point was the easy part! Now I just needed to figure out how to get the data into the AIPS++ system. The way I am currently doing this is by using the tablefromascii() command in AIPS++. I will give you an example of what I mean here in a few seconds. This command allows the AIPS++ system to import an ASCII file into a table. Now as promised here is a quick example of the way I plot my data in AIPS++:
-include pgplotter.g ; Include the pgplotter program
-t:=tablefromascii( 01230103', /home/pw/01230103.LBR ,,T);
This line tells AIPS++ to import the ASCII file 01230103.LBR
into a table named 01230103.
-x:=t.getcol( Column1'); Let x = the first column in the
file number of seconds since
00:00 LST.
-y:=t.getcol( Column2'); Let y = the value read from the
dish.
-pg:=pgplotter(); Let pg = the pgplotter function
and open the plotting window.
-pg.plotxy(x,y); Plot time as x axis and readings
as the y axis.
From here you can add additional plots to the same window by
doing the following command:
-pg.plotxy(x,y1,,newplot=F) Where y1 is the read value from
the dish for another observation.
Well as you can see I am still way down on the food chain as far as the AIPS++ system goes, but I felt that if I wrote this and any other Amateur read it that they would be able to do the same thing that I have done. Since I first started using the AIPS++ system I have added yet another dish to my setup. As I write this I am attempting to learn how I would be able to do synthesis with the two dishes. That way they will seem like one dish 28' (8.4 meter) dish. I am also in the process of building an audio detector for my setup with the hopes of being able to record the audio data to a HI-FI VCR for late play back an spectrum analysis. As I learn more about the AIPS++ system, I will submit more articles to the newsletter for the other Amateurs out there that are wishing that they could do the same things as "The Big Boys".
|