|
|
|||
| NRAO Home > CASA > CASA Cookbook and User Reference Manual |
|
||
8.3.11 Single Dish Spectral Analysis Use Case With ASAP Toolkit
Below is a script that illustrates how to reduce single dish data using ASAP within CASA. First a summary of the dataset is given and then the script.
#
# Project: AGBT06A_018_01
# Observation: GBT(1 antennas)
#
#Data records: 256 Total integration time = 1523.13 seconds
# Observed from 01:45:58 to 02:11:21
#
#Fields: 4
# ID Name Right Ascension Declination Epoch
# 0 OrionS 05:15:13.45 -05.24.08.20 J2000
# 1 OrionS 05:35:13.45 -05.24.08.20 J2000
# 2 OrionS 05:15:13.45 -05.24.08.20 J2000
# 3 OrionS 05:35:13.45 -05.24.08.20 J2000
#
#Spectral Windows: (8 unique spectral windows and 1 unique polarization setups)
# SpwID #Chans Frame Ch1(MHz) Resoln(kHz) TotBW(kHz) Ref(MHz) Corrs
# 0 8192 LSRK 45464.3506 6.10423298 50005.8766 45489.3536 RR LL HC3N
# 1 8192 LSRK 45275.7825 6.10423298 50005.8766 45300.7854 RR LL HN15CO
# 2 8192 LSRK 44049.9264 6.10423298 50005.8766 44074.9293 RR LL CH3OH
# 3 8192 LSRK 44141.2121 6.10423298 50005.8766 44166.2151 RR LL HCCC15N
# 12 8192 LSRK 43937.1232 6.10423356 50005.8813 43962.1261 RR LL HNCO
# 13 8192 LSRK 42620.4173 6.10423356 50005.8813 42645.4203 RR LL H15NCO
# 14 8192 LSRK 41569.9768 6.10423356 50005.8813 41594.9797 RR LL HNC18O
# 15 8192 LSRK 43397.8198 6.10423356 50005.8813 43422.8227 RR LL SiO
# Scans: 21-24 Setup 1 HC3N et al
# Scans: 25-28 Setup 2 SiO et al
casapath=os.environ[’AIPSPATH’]
#ASAP script # COMMENTS
#-------------------------------------- -----------------------------------------------
import asap as sd #import ASAP package into CASA
#Orion-S (SiO line reduction only)
#Notes:
#scan numbers (zero-based) as compared to GBTIDL
#changes made to get to OrionS_rawACSmod
#modifications to label sig/ref positions
os.environ[’AIPSPATH’]=casapath #set this environment variable back - ASAP changes it
s=sd.scantable(’OrionS_rawACSmod’,False)#load the data without averaging
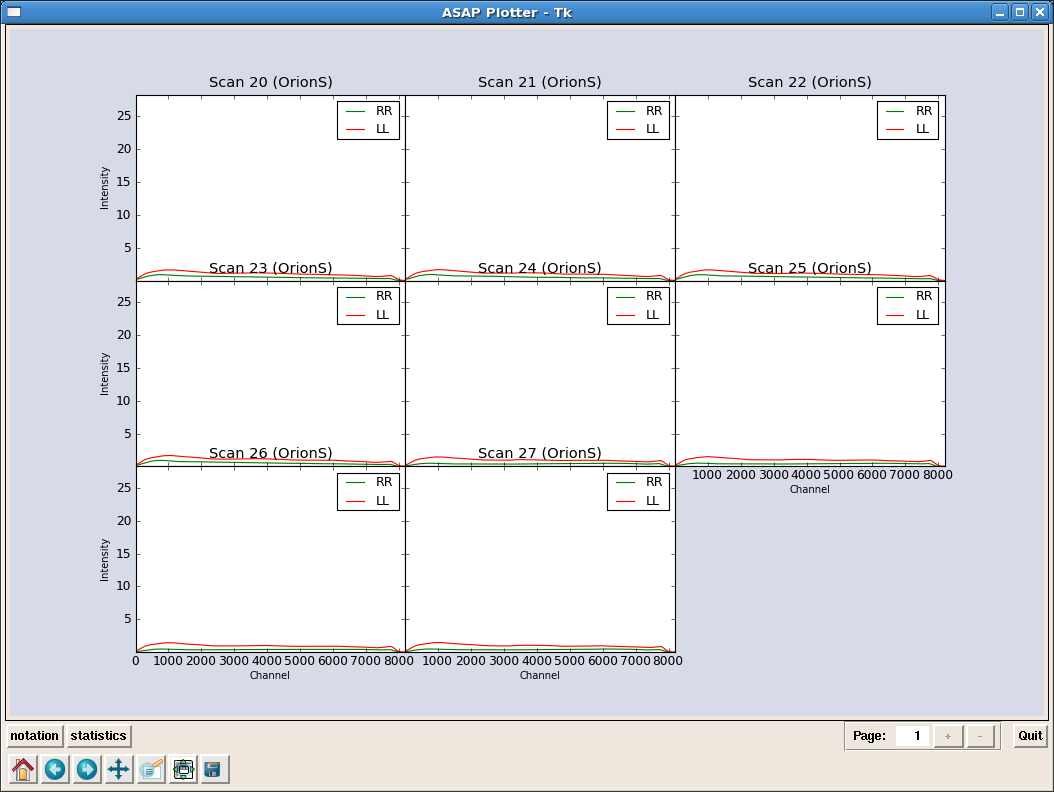
_________________________________________________________________________________________
s.set_fluxunit(’K’) # make ’K’ default unit
scal=sd.calps(s,[20,21,22,23]) # Calibrate HC3N scans
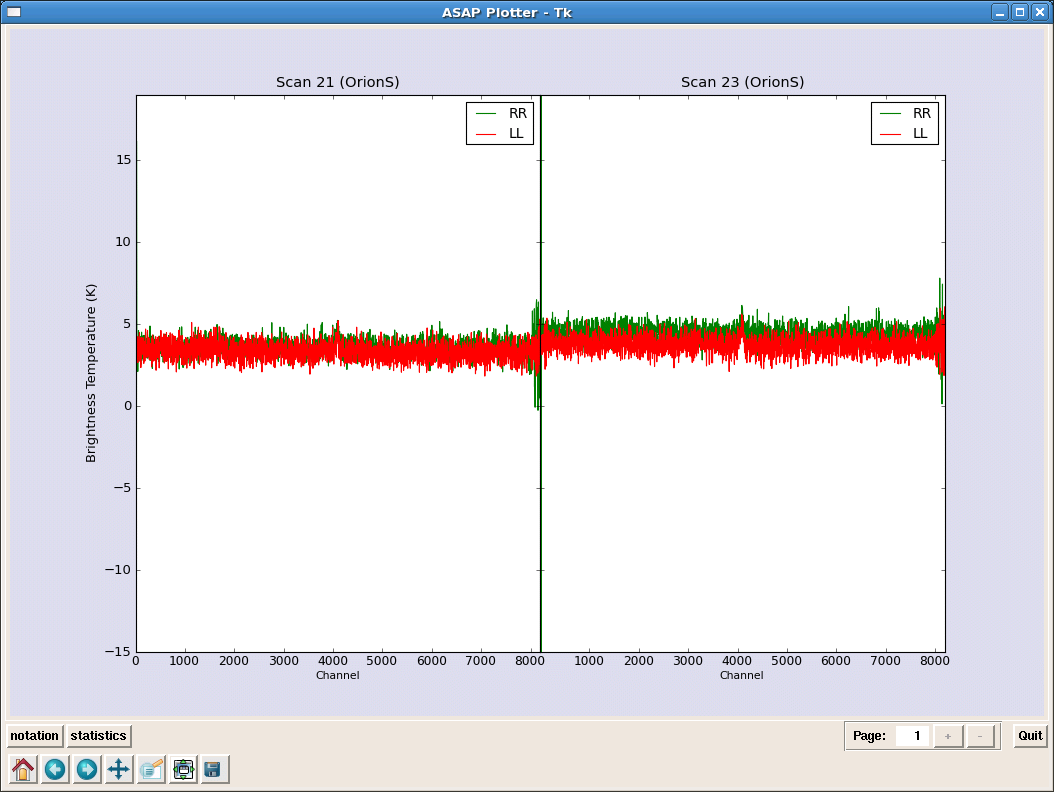
_________________________________________________________________________________________
scal.opacity(0.09) # do opacity correction
sel=sd.selector() # Prepare a selection
sel.set_ifs(0) # select HC3N IF
scal.set_selection(sel) # get this IF
stave=sd.average_time(scal,weight=’tintsys’) # average in time
spave=stave.average_pol(weight=’tsys’) # average polarizations;Tsys-weighted (1/Tsys**2) average
sd.plotter.plot(spave) # plot
spave.smooth(’boxcar’,5) # boxcar 5
spave.auto_poly_baseline(order=2) # baseline fit order=2
sd.plotter.plot(spave) # plot
spave.set_unit(’GHz’)
sd.plotter.plot(spave)
sd.plotter.set_histogram(hist=True) # draw spectrum using histogram
sd.plotter.axhline(color=’r’,linewidth=2) # zline
sd.plotter.save(’orions_hc3n_reduced.eps’)# save postscript spectrum
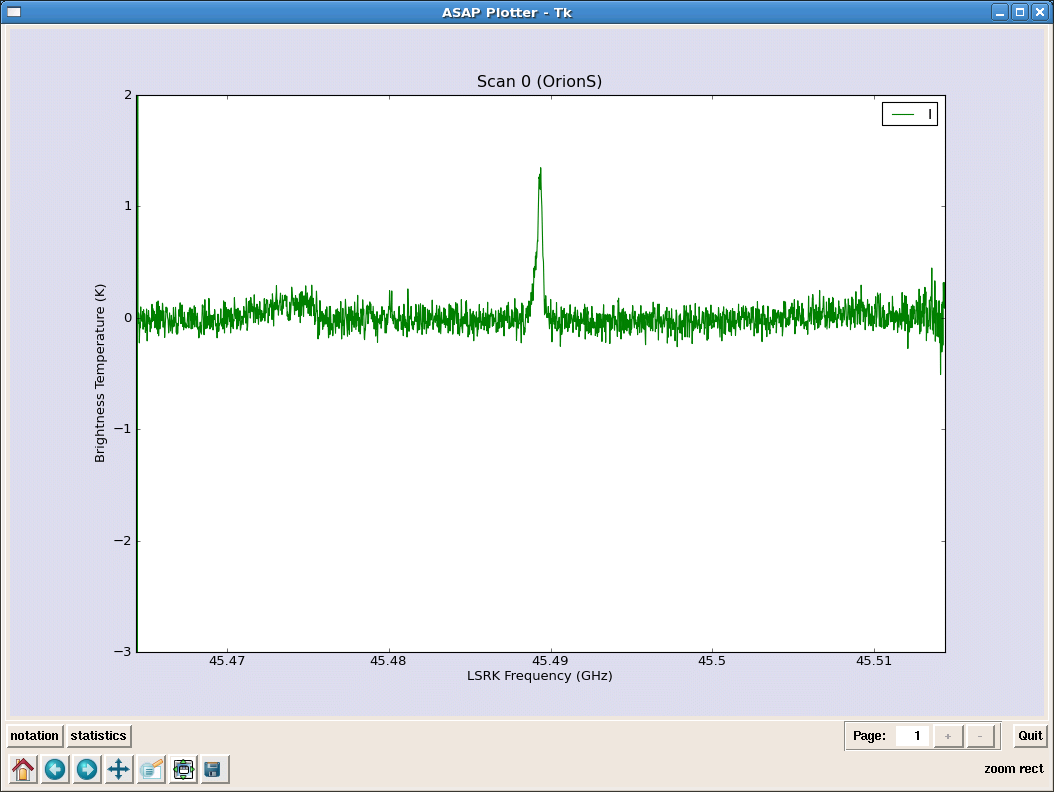
_________________________________________________________________________________________
rmsmask=spave.create_mask([5000,7000]) # get rms of line free regions
rms=spave.stats(stat=’rms’,mask=rmsmask)# rms
#----------------------------------------------
#Scan[0] (OrionS_ps) Time[2006/01/19/01:52:05]:
# IF[0] = 0.048
#----------------------------------------------
# LINE
linemask=spave.create_mask([3900,4200])
max=spave.stats(’max’,linemask) # IF[0] = 0.918
sum=spave.stats(’sum’,linemask) # IF[0] = 64.994
median=spave.stats(’median’,linemask) # IF[0] = 0.091
mean=spave.stats(’mean’,linemask) # IF[0] = 0.210
# Fitting
spave.set_unit(’channel’) # set units to channel
sd.plotter.plot(spave) # plot spectrum
f=sd.fitter()
msk=spave.create_mask([3928,4255]) # create region around line
f.set_function(gauss=1) # set a single gaussian component
f.set_scan(spave,msk) # set the data and region for the fitter
f.fit() # fit
f.plot(residual=True) # plot residual
# 0: peak = 0.786 K , centre = 4091.236 channel, FWHM = 70.586 channel
# area = 59.473 K channel
f.store_fit(’orions_hc3n_fit.txt’) # store fit
# Save the spectrum
spave.save(’orions_hc3n_reduced’,’ASCII’,True) # save the spectrum
More information about CASA may be found at the
CASA web page
Copyright © 2010 Associated Universities Inc., Washington, D.C.
This code is available under the terms of the GNU General Public Lincense
Home |
Contact Us |
Directories |
Site Map |
Help |
Privacy Policy |
Search
