clean(algorithm, niter, gain, threshold, displayprogress, model, fixed, complist, mask, image, residual, interactive, npercycle, masktemplate, async)
Description
Makes a clean image using either the Hogbom, Clark, multi-scale or multi-field
algorithms. The Clark algorithm is the default. The clean is performed
on the residual image calculated from the visibility data currently
selected. Hence the first step performed in clean is to transform the
current model or models (optionally including a componentlist) to fill
in the MODEL_DATA column, and then inverse transform the residual
visibilities to get a residual image. This residual image is then
cleaned using the corresponding point spread function. This means that
the initial model is used as the starting point for the
deconvolution. Thus if you want to restart a clean, simply set the
model to the model that was previously produced by clean.
Rather than explicit CLEAN boxes, mask images are used to constrain
the region that is to be deconvolved. To make mask images,
use either boxmask (to define a mask
via the corner locations blc and trc) or
mask (to define a mask via
thresholding an existing image) or regionmask (to make masks via regions using the regionmanager or interactively through the viewer) . The default mask is the inner quarter
of the image.
The CLEAN deconvolution is joint in whatever Stokes parameters are
present. Thus it searchs for peaks in either I or I + | V| or
I + 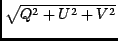 , the rationale for the latter two forms being
to be biased towards finding strongly polarized pixels first (these
forms are also the maximum eigenvalue of the coherency matrix). The
PSF is constrained to be the same in all polarizations (a feature of
this implementation, not of the Hamaker-Bregman-Sault formalism).
, the rationale for the latter two forms being
to be biased towards finding strongly polarized pixels first (these
forms are also the maximum eigenvalue of the coherency matrix). The
PSF is constrained to be the same in all polarizations (a feature of
this implementation, not of the Hamaker-Bregman-Sault formalism).
The clean algorithms possible are:
- Hogbom
- The classic algorithm: points are found iteratively
by searching for the peak. Each point is subtracted from the
full residual image using the shifted and scaled point spread
function.
- Multiscale
- An experimental multi-scale clean algorithm is invoked.
The algorithm is fully described in
deconvolver.
- Clark
- The faster algorithm: the cleaning is split into
minor and major cycles. In the minor cycles only the brightest
points are cleaned, using a subset of the point spread function.
In the major cycle, the points thus found are subtracted correctly
by using an FFT-based convolution.
- Multi-field
- Cleaning is split into minor and major
cycles. For each field, a Clark-style minor cycle is performed.
In the major cycle, the points thus found are subtracted
either from the original visibilities (for multiple fields)
or using a convolution (for only one field). The latter is
much faster. Multi-field imaging has been implemented for
Clark, Hogbom, and Multi-scale deconvolution algorithms.
- Cotton-Schwab
- Cleaning is split into minor and major
cycles. For each field, a Clark-style minor cycle is performed.
In the major cycle, the points thus found are subtracted
from the original visibilities. A fast variant does a convolution
using a FFT. This will be faster for large numbers of
visibilities. Double the image size from that used for Cotton-Schwab
and set a mask to clean only the inner quarter.
- Wide-field
- The user will need to use a wide-field algorithm to
deconvolve if the array is not coplanar over the field of view being
imaged . The technique used is to break the field being imaged into
smaller pieces (facets), over each of which the array appear
planar. We implement a rectangular facetting scheme. If the number of
facets specified in setimage is
greater than one, Either wfhogbom or wfclark algorithm has to be
selected here to perform a wide-field decovolution. The function
advise can be used to calculate or
check if you need to use a wide-field deconvolution. Note that
aliasing can be reduced by using the padding argument in
setoptions. In practice the
previous sentence means that if you notice the clean to diverge at the
edges of the facets then you need to use a larger amount of padding
for the FT; the default being 1.2. Wide-field imaging has been
implemented for Clark and Hogbom algorithms.
The multi-field clean should be used if either of two conditions
hold:
- 1.
- Multiple fields are to be cleaned simultaneously OR
- 2.
- Primary beam correction is enabled. In this case, a
mosaiced clean is performed.
Note that for the single pointing algorithms, only a quarter of the
image may be cleaned. If no mask is set, then the cleaned region
defaults to the inner quarter. If a mask larger than a quarter of the
image is set, then only the inner quarter part of that mask is used.
However, for the wide-field and multi-field imaging (including the
Cotton-Schwab algorithm), the entire field may be imaged because the
major cycles either do an exact subtraction from the visibilities or
because PSF extent is more than twice the extent of the primary beam
support.
Before clean can be run, you must run setdata and setimage.
Before clean can be run with a multi-field algorithm, you should run
setvp. You may want to run setmfcontrol before running clean
with a multi-field or wide-field algorithm, though the default control values
may be acceptable. Before clean can be run with a multi-scale algorithm,
setscales must be run.
Interactive cleaning/masking: If the user wants to see what the clean
image looks like after npercycle iteration and mask or modify the mask
each time, he/she should set interactive=T and give npercycle to a
fraction of niter. A viewer with the last residual image along with an
overlayed mask appear after every npercycle iteration. The user can
add or delete regions (by clicking on the appropriate button) to the
mask using the region button and drawing regions and double clicking
inside the region. Use 'refresh mask' to review the recent changes in
the mask. When satisfied and ready to continue cleaning press 'DONE
with masking' (if the user want to terminate the cleaning process use
the 'STOP' button). The button 'No more mask changes' should be used
if the user want clean to proceed without any further interruption.
PLEASE be patient and wait for the viewer to load both the image and
mask before clicking on the buttons or drawing regions. If impatient
please use interactive=F. Even if interactive=F, and if the parameter
'mask' is non-empty, it is still used in limiting the search area for
clean components.
If the parameter 'masktemplate' is not empty this means that the user
want to use a prior image to make the mask the first time (e.g
a previously cleaned image)
Arguments
| algorithm |
|
Algorithm to use |
|
|
Allowed: |
String:'clark'|'hogbom'|'multiscale'|'mfclark'|'csclean'|'csfast'|
'mfhogbom'|'mfmultiscale'|'wfclark'|'wfhogbom' |
|
|
Default: |
'clark' |
| niter |
|
Number of Iterations, set to zero for no CLEANing |
|
|
Allowed: |
Int |
|
|
Default: |
1000 |
| gain |
|
Loop Gain for CLEANing |
|
|
Allowed: |
Float |
|
|
Default: |
0.1 |
| threshold |
|
Flux level at which to stop CLEANing |
|
|
Allowed: |
Quantity |
|
|
Default: |
'0Jy' |
| displayprogress |
|
Display the progress of the cleaning? |
|
|
Allowed: |
Bool |
|
|
Default: |
F |
| model |
|
Names of clean model images |
|
|
Allowed: |
Vector of strings |
| fixed |
|
Keep one or more models fixed |
|
|
Allowed: |
Vector of booleans |
|
|
Default: |
F |
| complist |
|
Name of component list |
|
|
Allowed: |
String |
| mask |
|
Names of mask images used for CLEANing |
|
|
Allowed: |
Vector of strings |
| image |
|
Names of restored images |
|
|
Allowed: |
Vector of strings |
| residual |
|
Names of residual images |
|
|
Allowed: |
Vector of strings |
| interactive |
|
whether to stop clean and interactively mask |
|
|
Allowed: |
Boolean |
|
|
Default: |
F |
| npercycle |
|
If interactive is 'T', then no of iter of clean
before stopping, usually a fraction of niter |
|
|
Allowed: |
Int |
|
|
Default: |
100 |
| masktemplate |
|
If non empty then will use this image to make
the mask the first time |
|
|
Allowed: |
String |
|
|
Default: |
'' |
| async |
|
Run asynchronously in the background? |
|
|
Allowed: |
Bool |
|
|
Default: |
!dowait |
Returns
Bool
Example
imgr.clean(model='3C273XC1.clean.model',
mask='3C283XC1.mask', niter=1000, gain=0.25, threshold='0.03Jy')
A few points should be noted in this example:
- When the mask parameter is specified, the number of mask images
listed should be equal to the number of model images. They
should also have the same coordinate system as their
corresponding model images.
- If one or more model images are listed in the model parameter
but the image and residual parameters are empty, the restored
and residual images are automatically named as the model names
appended with '.restored' and '.residual', respectively.
- No restored image is made if the image string is explicitly unset.
include 'qimager.g';
msfile := 'vlac125K.ms';
imgr:=qimager(msfile);
npix := 500; cell:='5arcsec';
#
# CS on 500 by 500
#
imgr.setimage(nx=npix, ny=npix, cellx=cell, celly=cell, stokes='I',
spwid=[1,2]);i
imgr.setoptions(padding=1.0);
imgr.setdata(spwid=[1,2]);
imgr.clean('cs', model='vlac125K.cs', image='vlac125K.cs.restored',
niter=1000, gain=0.1);
#
# CSF on 1000 by 1000, cleaning inner quarter
#
include 'regionmanager.g';
imgr.setimage(nx=2*npix, ny=2*npix, cellx=cell, celly=cell, stokes='I',
spwid=[1,2]);
imgr.regionmask('vlac125K.mask', region=drm.quarter());
imgr.clean('csf', model='vlac125K.csf', image='vlac125K.csf.restored',
mask='vlac125K.mask', niter=1000, gain=0.1);
#
# CS on 1000 by 1000, cleaning entire image
#
include 'regionmanager.g';
imgr.setimage(nx=2*npix, ny=2*npix, cellx=cell, celly=cell, stokes='I',
spwid=[1,2]);
imgr.clean('cs', model='vlac125K.csl', image='vlac125K.csl.restored',
mask='vlac125K.mask', niter=1000, gain=0.1);
imgr.done();



 Next: qimager.mem - Function
Up: qimager - Tool
Previous: qimager.setbeam - Function
Contents
Index
Please send questions or comments about AIPS++ to aips2-request@nrao.edu.
Next: qimager.mem - Function
Up: qimager - Tool
Previous: qimager.setbeam - Function
Contents
Index
Please send questions or comments about AIPS++ to aips2-request@nrao.edu.
Copyright © 1995-2000 Associated Universities Inc.,
Washington, D.C.
Return to AIPS++ Home Page
2006-10-15

 News
News  FAQ
FAQ
 Search
Search
 Home
Home

![]() , the rationale for the latter two forms being
to be biased towards finding strongly polarized pixels first (these
forms are also the maximum eigenvalue of the coherency matrix). The
PSF is constrained to be the same in all polarizations (a feature of
this implementation, not of the Hamaker-Bregman-Sault formalism).
, the rationale for the latter two forms being
to be biased towards finding strongly polarized pixels first (these
forms are also the maximum eigenvalue of the coherency matrix). The
PSF is constrained to be the same in all polarizations (a feature of
this implementation, not of the Hamaker-Bregman-Sault formalism).
![]()
![]()
![]()
![]()
![]()