
 News News |
 FAQ FAQ
|
 Search Search
|
 Home Home
|
| Getting Started | Documentation | Glish | Learn More | Programming | Contact Us |

| Version 1.9 Build 1488 |
|
| Package | general | |
| Module | images | |
| Tool | image |
| moments | in | List of moments that you would like to compute | |
| Allowed: | Vector of integers | ||
| Default: | 0 (integrated spectrum) | ||
| axis | in | The moment axis | |
| Allowed: | Positive integer | ||
| Default: | The spectral axis if there is one | ||
| region | in | Region of interest | |
| Allowed: | Region tool | ||
| Default: | Whole image | ||
| mask | in | OTF mask | |
| Allowed: | Boolean LEL expression or mask region | ||
| Default: | None | ||
| method | in | List of windowing and/or fitting functions you would like to invoke | |
| Allowed: | Vector of strings from `window', `fit', and `interactive' | ||
| Default: | Don't invoke the window or fit functions, and don't invoke any interactive functions | ||
| smoothaxes | in | List of axes to smooth | |
| Allowed: | Vector of integers | ||
| Default: | No smoothing | ||
| smoothtypes | in | List of smoothing kernel types, one for each axis to smooth | |
| Allowed: | Vector of strings from `gauss', `boxcar', `hanning' | ||
| Default: | No smoothing | ||
| smoothwidths | in | List of widths (full width for boxcar, full width at half maximum for gaussian, 3 for Hanning) in pixels for the smoothing kernels | |
| Allowed: | Vector of numeric, quantity or string | ||
| Default: | No smoothing | ||
| includepix | in | Range of pixel values to include | |
| Allowed: | Vector of 1 or 2 Floats | ||
| Default: | Include all pixels | ||
| excludepix | in | Range of pixel values to exclude | |
| Allowed: | Vector of 1 of 2 Floats | ||
| Default: | Exclude no pixels | ||
| peaksnr | in | The SNR ratio below which the spectrum will be rejected as noise (used by the window and fit functions only) | |
| Allowed: | A positive float | ||
| Default: | 3 | ||
| stddev | in | Standard deviation of the noise signal in the image (used by the window and fit functions only) | |
| Allowed: | Positive float | ||
| Default: | Automatic noise determination | ||
| doppler | in | Velocity doppler definition for velocity computations along spectral axes | |
| Allowed: | String | ||
| Default: | RADIO | ||
| outfile | in | Output image file name (or root for multiple moments) | |
| Allowed: | String | ||
| Default: | input + an auto-determined suffix | ||
| smoothout | in | Output file name for convolved image | |
| Allowed: | String | ||
| Default: | Don't save the convolved image | ||
| plotter | in | The PGPLOT device name to make plots on | |
| Allowed: | Any valid PGPLOT device | ||
| Default: | No plotting | ||
| nx | in | The number of subplots per page in the x direction | |
| Allowed: | Any positive integer | ||
| Default: | 1 | ||
| ny | in | The number of subplots per page in the y direction | |
| Allowed: | Any positive integer | ||
| Default: | 1 | ||
| yind | in | Scale the y axis of the profile plots independently | |
| Allowed: | Bool | ||
| Default: | False | ||
| overwrite | in | Overwrite (unprompted) pre-existing output file ? | |
| Allowed: | T or F | ||
| Default: | F | ||
| drop | in | Drop moments axis from output images ? | |
| Allowed: | T or F | ||
| Default: | T | ||
| async | in | Run asynchronously? | |
| Allowed: | Bool | ||
| Default: | !dowait - but always F if plotting | ||
Summary
The primary goal of this function is to enable you to analyze a multi-dimensional image by generating moments of a specified axis. This is a time-honoured spectral-line analysis technique used for extracting information about spectral lines. There is also a custom GUI interface available via the momentsgui function as the command line interface is presently rather cumbersome.
You can generate one or more output moment images. The return value of this function is an Image tool holding the first of the output moment images.
The word `moment' is used loosely here. It refers to collapsing an axis (the moment axis) to one pixel and setting the value of that pixel (for all of the other non-collapsed axes) to something computed from the data values along the moment axis. For example, take an RA-DEC-Velocity cube, collapse the velocity axis by computing the mean intensity at each RA-DEC pixel. This function offers many different moments and a variety of interactive and automatic methods to compute them.
We try to make a distinction between a `moment' and a `method'. This boundary is a little blurred, but it claims to refer to the distinction between what you are computing, and how the pixels that were included in that computation were selected. For example, a `moment' would be the average value of some pixel values in a spectrum. A `method' for selecting those pixels would be a simple pixel value range specifying which pixels should be included.
There are many available moments, and you specify each one with an integer code as it would get rather cumbersome to refer to them via strings. In the list below, the value of the ith pixel of the spectrum is Ii, the coordinate of this pixel is vi (of course it may not be velocity), and there are n pixels in the spectrum. The available moments are:
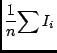
where ![]() v is the width (in world coordinate units) of a pixel
along the moment axis
v is the width (in world coordinate units) of a pixel
along the moment axis
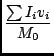
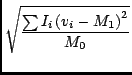
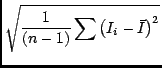
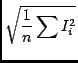
Smoothing
The purpose of the smoothing functionality is purely to provide a mask. Thus, you can smooth the input image, apply a pixel include or exclude range, and generate a smoothed mask which is then applied before the moments are generated. The smoothed data are not used to compute the actual moments; that is always done from the original data.
Basic Method
The basic method is to just compute moments directly from the pixel
values. This can be modified by applying pixel value inclusion or
exclusion ranges (arguments includepix and excludepix).
You can then also convolve the image (arguments smoothaxes, smoothtypes, and smoothwidths) and find a mask based on the inclusion or exclusion ranges applied to the convolved image. This mask is then applied to the unsmoothed data for moment computation.
Window Method
The window method (invoked with argument method='window') does no pixel-value-based selection. Instead a window is found (hopefully surrounding the spectral line feature) and only the pixels in that window are used for computation. This window can be found from the convolved or unconvolved image (arguments smoothaxes, smoothtypes, and smoothwidths).
The moments are always computed from the unconvolved data. The window
can be found (for each spectrum) interactively or automatically. The
automatic methods are via Bosma's converging mean algorithm (method='window') or by fitting Gaussians and taking
![]() 3
3![]() as
the window (method='window,fit'). The interactive methods
are a direct specification of the window with the cursor (method='window,interactive') and by interactive fitting of Gaussians
and taking
as
the window (method='window,fit'). The interactive methods
are a direct specification of the window with the cursor (method='window,interactive') and by interactive fitting of Gaussians
and taking ![]() 3-sigma as the window (method='window,fit,interactive').
3-sigma as the window (method='window,fit,interactive').
In Bosma's algorithm, an initial guess for a range of pixels surrounding a spectral feature is refined by widening until the mean of the pixels outside of the range converges (to the noise).
Fit Method
The fit method (method='fit') fits Gaussians to spectral features either automatically or interactively (method='fit,interactive'). The moments are then computed from the Gaussian fits (not the data themselves).
The interactive methods are very user intensive. You have to do something for each spectrum. These are really only useful for images with a manageably small number of spectra.
Other Arguments
Note also that if you specify a plotting device but do not ask for an interactive option, then plots will be made showing you the effect of the algorithm on each spectrum. For example, you may have selected the automatic windowing method. If you have specified a plotting device, then each spectrum will be plotted and the window marked on each plot.
The convolving point-spread function is normalized to have a volume of unity. This means that point sources are depressed in value, but extended sources that are large with respect to the PSF remain essentially on the same intensity scale; these are the structures you are trying to find with the convolution so this is what you want. If you convolve the image, then arguments like includepix select based upon the convolved image pixel values. If you are having trouble getting these right, you can output the convolved image (smoothout) and assess the validity of your pixel ranges. Note also that if you are Hanning convolving (usually used on a velocity axis), then the width for this kernel must be 3 pixels (triangular smoothing kernels of other widths have no valid theoretical basis).
Finally, if you ask for a moment which requires the coordinate to be computed for each profile pixel (these are the intensity weighted mean coordinate [moment 1] and the intensity weighted dispersion of the coordinate [moment 2]), and the profile axis is not separable then there will be a performance loss. Examples of non-separable axes are RA and Dec. If the axis is separable (e.g. a spectral axis) there is no penalty. In the latter case, the vector of coordinates for one profile is the same as the vector for another profile, and it can be precomputed (once).
Note that this function has no ``virtual'' output file capability. All output files are written to disk. The output mask for these images is good (T) unless the moment method fails to generate a value (e.g. the total input pixel mask was all bad for the profile) in which case it will be bad (F).
- im2 := im.moments(moments=[-1,1,2],axis=3,smoothaxes=[1,2,3],
smoothtypes="gauss gauss hann",smoothwidths=[5.0,5.0,3],
excludepix=[1e-3], smoothout='smooth')
In this example, standard moments (average intensity, weighted velocity and weighted velocity dispersion) are computed via the convolve (spatially convolved by gaussians and spectrally by a Hanning kernel) and clip method (we exclude any pixels with absolute value less than 0.001). The output file names are automatically created for us and the convolved image is saved. The returned image tool holds the first moment image.
- im2 := im.moments(moments=[3],method='window',plotter='/glish',
nx=5,ny=5)
In this example, the median of each spectrum is computed, after pixel selection by the automatic window method. Each spectrum and the window are plotted (25 plots per page) on a PGPLOT X-window device. Because the plotting device is given and we did not give the noise level of the image, a histogram of the image is made and fit and we get to interactively do the Gaussian fit to the histogram. The output image is temporary and accessed via the returned Image tool.