
 News News |
 FAQ FAQ
|
 Search Search
|
 Home Home
|
| Getting Started | Documentation | Glish | Learn More | Programming | Contact Us |

| Version 1.8 Build 235 |
|
J McMullin, J Braatz, B Garwood
This chapter provides brief guidelines on how to reduce the currently supported observing modes at the GBT.
An example data reduction session is shown to demonstrate the look and feel of data reduction in DISH. We calibrate and average position switched scans.
> . /home/aips++/gbt/aipsinit.sh (BASH)
> source /home/aips++/gbt/aipsinit.csh (T/CSH)
>
> dish
- d.open('stjul13SP')
- d.gms()
Scan Object Proctype SWState SWtchsig Procseqn Procsize
35 S8 Track FSWITCH FSW01 1 1
36 S8 Track FSWITCH FSW12 1 1
37 S8 Track FSWITCH FSW12 1 1
38 S8 OffOn PSWITCHOFF TPWCALSP 1 2
40 S8 OnOff PSWITCHON TPWCALSP 1 2
42 S8 Track NONE TPWCALSP 1 1
- d.calib(36)
- d.calib(37)
- avg\_spc:=d.aver([36,37]);
- d.plotscan(avg\_spc)
- d.header()
Proj : standards\_jul13 Src : S8 Proc : Track
Obs : Richard RA : 05:47:21.406 PType: SIMPLE
Scan : 36 Dec : -001.40.19.036 OType: LINE
Seq : 1/1 Epoch: J2000 Swtch: FSWITCH
Date : 2002-07-13 Az : -112.772 Swsig: FSW12
Time : 19:31:14.500 UT El : 23.733 Ints : 31
Tsys : 16.58 Trx : 7.58 Tcal : 1.60
Tsys : 16.86 Trx : 8.88 Tcal : 1.64
BW : 4.995 (MHz) Res : 4.883 (kHz)
Expos: 54.549 Durat: 247.856
T
- d.setregion()
Use the left button to set location, right button to exit
region = [15 180 296 681 810 1009]
T
- d.nfit(1)
T
- d.bshape()
T
- d.fileout('S8\_avgbl')
New file S8\_avgbl is created
T
- d.keep()
globalscan saved to S8\_avgbl
T
The following lists the most often used utilities. There are many more available functions in dish. Type "help('Refman:dish')" at the command line prompt to redirect your browser to the DISH command line documentation; 'd.info()' will also provide an alphabetical, table of all available DISH functions); typing d.help('function_name') will provide information on the function (d.help('function_name',T) will also automatically drive your web browser to the AIPS++ User Reference Manual entry on this function).
| File Access | Scan Access | Editing | Calibration | Analysis | Display |
| import | getr | msplot | calib | (all operations) | plotr |
| open | getc | viewer | TPcal | e.g., aver | plotc |
| filein | getscan | clip | FScal | base | plotscan |
| fileout | flag | SRcal | etc |
GBT data must be converted from their raw FITS files to an AIPS++ MeasurementSet before being accessed with dish functions. The command to fill raw GBT data is import. Once data have been filled, they can be accessed with the open function. Two additional file access functions, filein and fileout, are used to set the focus for where data is read from and written to.
![]()
Example:
d.import(projdir='/home/gbtdata/OHsurvey',outms='oh',startscan=100,stopscan=105)
Parameters:
Example:
d.open('ohSP');
Parameters:
Example:
d.filein('ohSP')
Parameters:
Example:
d.fileout('avg_spectra')
Parameters:
Data in the MeasurementSet contains both the uncalibrated (raw or observed data) and the calibrated data. Initially, the calibrated data is simply a copy of the uncalibrated data. As the data is manipulated through the calibration routines (calib, TPcal, FScal, SRcal), the calibrated data becomes distinct.
The SDRecord (single dish record) is the data structure returned by the data access commands. The basic SDRecord has four sub-records which contain basic information on the observation:
Several dish functions operate on a globally accessible SDRecord (spectrum) to help simplify the interface. In general, these functions will operate on the currently displayed spectrum or the contents of this globally accessible record (called globalscan1). Functions which take no arguments will in general access this spectrum and perform the operation on it.
Example:
- s35_1 := d.getr(35,1,1,1) #retrieve scan 35, phase 1, integration 1,
#polarization 1 from the opened MS and
#store in variable s35_1
- s35_1 := d.getr(35,1) #retrieve scan 35, phase 1, average all
#integrations, show all polarizations
Parameters:
Example:
- s35_c1 := d.getc(35,1,1) #retrieve scan 35, integration 1,
#polarization 1 from the opened MS and
#store in variable s35_c1
- s35_c1 := d.getr(35) #retrieve scan 35, average all integrations,
#show all polarizations
Parameters:
Example:
- s35_c1 := d.getscan(35,3) #retrieve scan 35, subscan 1
Generally, you need to understand how the data was taken to effectively use getscan.
Parameters:
![]()
Flagging data is done in two basic modes: 1) command line and 2) GUI. There are two basic command line flagging utilities, clip and flag.
clip is a simple function which will allow you to flag a currently viewed spectrum.
Example:
d.clip(0.4); #clip all above 0.4 d.clip(0.4,[100:500]); #clip above 0.4 in the channel range 100-500 d.clip(-0.3,clipdir='low'); #clip below -0.3
Parameters:
flag is a more complete flagging utility allowing versatile flagging of data. Automated flagging based on data statistics is under development for single dish data.
Example:
d.open('stjul13SP');
d.gms();
d.plotc(36)
d.flag(36,channel=100:120) #flag all subscans, polarizations of scan
#36 in the channel range of 100:120
d.plotc(36) #you should see the range masked
d.flag(36,flag=F); #unset all flagging
d.plotc(36) #it's back!
d.flag(36,channel=500:550,polarization=1)
d.plotc(36) #you should see a gap just in the first
#polarization
d.flag(36,flag=F);
d.flag(36,1) #flag just the first integration
d.plotc(36) #should look okay though it doesn't
#include the first integration
d.plotc(36,1) #masked
d.plotc(36,2) #okay
Parameters:
autoflag is a utility within AIPS++ which allows automated (based on statistics, etc) flagging of data without direct examination of each flagged row/scan. A few specific examples are provided below. See the
Flag based on statistics
include 'autoflag.g'
af:=autoflag('stjul13SP') #Specify the MeasurementSet to operate on
af.setfreqmed(); #Use a sliding median filter in frequency
# default threshold is 5
af.run(plotscr=F);
af.done(); #important to release the MeasurementSet
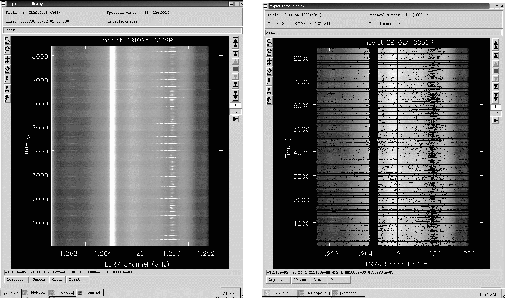 |
Clip data using autoflag
include 'autoflag.g'
af:=autoflag('stjul13SP') #Specify the MeasurementSet to operate on
cliprec:=[=] #set up a record to contain the clipping info
cliprec[1]:=[expr='ABS XX',min=0,max=200]
#clip the XX correlation above 200 and below 0
af.setselect(clip=cliprec) #specify the clip record as the selection
af.run(plotscr=F) #execute (disable intermediate displays)
GUI modes of flagging are also enabled through the viewer and msplot tools.
For the viewer:
- include 'viewer.g' - dv.gui();
This will bring up two GUIs. The first is for selecting what to display. You select the MeasurementSet and choose the type of display (only 'Raster' is available for MeasurementSets). This will then be displayed in the other display panel.
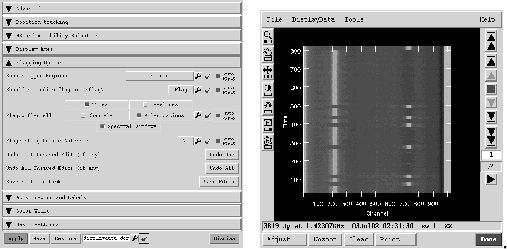 |
For msplot:
- include 'msplot.g'
- mp := msplot('your\_measurementset\_name');
This brings up a GUI which will allow you to select the type of display ("X vs. Y" or "Display data as image") and optionally select a subset of the data.
arameters:
Parameters:
Parameters:
The plotter is the standard dish display for spectral line data. It is built on the existing AIPS++ pgplotter tool and so has all of the basic PGPLOT functionality. In addition, the plotter features tools for:
Calibration is applied to the data based on information in the STATE table of the MS, in particular, it utilizes the OBSMODE column to determine a procedure type, switching signal, and switching state. In addition, information on the procedure size and procedure sequence number are used.
The function applies a number of corrections:
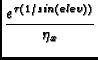
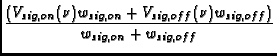
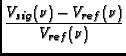
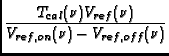 >
>
Parameters:
Parameters:
Parameters:
Parameters:
Each of the following operations has a GUI counterpart. We describe only the command line interface.
The functionality for averaging is provided in the aver function.
aver - Average a group of scans, subscans.
Example:
- myavg := d.aver([303,305:308]) #average scans 303,305,306,307,308 - myavg := d.aver([159:165],'1/4') #average every fourth subscan in - #scans 159-165
Generally, you need to understand how the data was taken to effectively use getscan.
Parameters:
There is also a means of averaging individual scans using an accumulator. Three functions are relevant: sclear(), accum(), and ave().
Example:
d.sclear() # clear the accumulator d.getc(35) # retrieve calibrated scan 35 d.accum() # add the scan to the accumulator d.getc(36) # retrieve calibrated scan 36 d.accum() # add the scan to the accumulator d.ave() # average scans in the accumulator d.show() # display the result on the plotter
Function sclear() takes no parameters, and clears the accumulator.
Function accum() also takes no parameters, and it adds the current contents of the globalscan1 spectrum into the accumulator.
Function ave() averages the contents of the accumulator, and the average is returned to the globalscan1 spectrum for further processing.
A UNIPOPS-like stack is also available. The stack is simply a vector of scan numbers which are intended to be averaged, or operated on in some other manner. It is possible to use Glish methods to accomplish the formation and manipulation of vector of scan numbers; nevertheless, these stack techniques provide another path for those familiar with the UNIPOPS syntax.
Four functions are used to manipulate the stack: empty(), addstack(), delete() and tellstack().
Example:
d.empty() # clear the stack d.addstack(10,50,2) # add even scans from 10 through 50 to the stack d.delete(24) # remove scan 24 d.delete(28) # remove scan 28 d.tellstack() # show the contents of the stack d.aver(d.tellstack()) # average the scans listed in the stack
Function empty() takes no parameters, and simply clears the stack.
Function addstack(beg,end=beg,inc=1) is used to append new entries to the stack. Parameters:
So, addstack(10) appends scan 10 to the stack; addstack(15,18) appends scans 15, 16, 17 and 18 to the stack, and addstack(21,28,3) appends scans 21, 24 and 27 to the stack. If these three commands were executed on an empty stack, the result would be [10, 15, 16, 17, 18, 21, 24, 27].
Function delete(value) removes a given scan from the stack. So to remove scan 16 from the stack constructed above, execute d.delete(16).
Function tellstack() takes no parameters, and is used to list the entries in the stack, and also to pass the stack to other functions in AIPS++. For example, to average the entries in the stack, execute d.aver(d.tellstack()).
Baselines can be removed during the calibration step; this is the most efficient means if the baseline regions are constant and there are many to perform. Additional functions for baselining are also provided in the base function.
base - Perform a polynomial or sinusoidal baseline fit to a scan.
Example:
mybl := d.base(order=2,action='subtract',range='[50:400],[1500:2000]')
# This will perform a 2nd order poly.
# fit to the currently displayed scan.
Generally, you need to understand how the data was taken to effectively use getscan.
Parameters:
One can also use fit a baseline with the UniPOPS-like functions.
d.nregion(50,100,900,1000)# Provide pairs of numbers as the channel range for the
# baseline region
d.setregion() # This will prompt the user to set the baseline regions
# with a cursor
d.nfit(1) # Fit a first order polynomial
d.bshape() # Show the baseline fit superposed on the spectrum
d.baseline() # Subtract the baseline; this also calls the
# d.rms() function displaying the baseline region stats
d.show() # Show the result
Fitting Gaussians is best done with the GUI interface. Pressing the GFit button on the dish plotter will pull up the Profile Fitter window (if there are multiple polarizations, they need to be fit separately and so should be first displayed with the getr or getc functions or after use of the getpol function).
The appropriate sequence of events is:
The Plotter will show the Estimate (yellow), the Fit (green) and the Residual (red). Each of these can be toggled on/off by going to the "Plot" menu in the Profile Fitter window and selecting/deselecting a plot.
The gauss fitting routine can also be used at the command line.
gauss - Fit gaussians to displayed spectrum.
Example:
- d.gauss(2) defaultregionmanager (drm) ready for use Creating (temp)image of shape [1023] Gauss: 1 Center: 1.42037e+00 Height: 5.44724e+01 Width: 4.20960e-05 Cerr: 3.71902e-07 Herr: 1.30446e+00 Werr: 1.20558e-06 Gauss: 2 Center: 1.42037e+00 Height: 9.25963e+00 Width: 1.71623e-04 Cerr: 4.43034e-06 Herr: 1.10727e+00 Werr: 1.43272e-05 T
Parameters:
There are several functions for obtaining statistical information on a given spectrum.
Example:
- d.stat() # look at spectrum (full range) [peak=68.9537735, area=0.00448239934, min=-0.303991884, rms=5.74451895, scan=35, centroid=1.42037562, vpeak=1.4203805, startint=1.41901819, stopint=1.42400843] - d.stat(range='[700:800]'); # look at line [peak=68.9537735, area=914.18058, min=0.242817193, rms=16.1565856, scan=35, centroid=745, vpeak=744, startint=700, stopint=800] - d.stat(range='[550:650]') # look at line free region [peak=0.180041969, area=-1.33219323, min=-0.202106252, rms=0.0901912276, scan=35, centroid=0, vpeak=554, startint=550, stopint=650]
Parameters:
This function will calculate statistics on the regions set via the setregion or nregion commands.
Example:
- d.rms() Using 1023 for calculating baseline stats. == Feed 1 === RMS = 3.24550665 Mean = 0.000737983237 Num points = 1023 max = 48.9361229 min = -12.9025164 nregion = [1 1023] == Feed 2 === RMS = 3.29074012 Mean = 0.351124813 Num points = 1023 max = 50.4430733 min = -14.0051394 nregion = [1 1023] T
This function will calculate statistics on a spectrum. It will use the global echan and bchan parameters for the region if available; these can also be set within the function.
Example:
- d.stats() Feed : 1 bchan: 1 rms : 5.744519 min : -0.303992 Npts : 1023 echan: 1023 mean : 0.898234 max : 68.953773 Feed : 2 bchan: 1 rms : 6.180395 min : -0.329462 Npts : 1023 echan: 1023 mean : 0.912738 max : 73.913177 T
Parameters:
This function will calculate statistics on a spectrum. It will use the global emoment and bmoment parameters for the region if available; these can be set with the bmoment and emoment functions.
Example:
- d.bmoment(700) T - d.emoment(800) T - d.moment() [peak=68.9537735, area=914.18058, min=0.242817193, rms=16.1565856, scan=35, centroid=745, vpeak=744, startint=700, stopint=800]
smooth - Smooth the resolution of a spectrum
Example:
- smooth_spc:=d.smooth(type='HANNING',decimate=T); - d.plotscan(smooth_spc)
Parameters:
boxcar, hanning, and chngres are streamlined cases for the smooth function using type BOXCAR, HANNING, or GAUSSIAN respectively, and with decimation=T.
boxcar - Perform a boxcar smooth on a spectrum
Example:
- d.show() - d.boxcar(5) - d.show()
Parameters:
hanning - Perform a hanning smooth on a spectrum
Example:
- d.show() - d.hanning() - d.show()
chngres - Perform a gaussian smooth on a spectrum
Example:
- d.show() - d.chngres(5) - d.show()
Parameters:
Saving spectra to into a new or existing output file is done using the fileout, lsoutfile, save, and keep commands. fileout was described in the Section on File access.
save, keep - Save spectrum to output file.
Example:
- d.files() Current filein is : N1SP Current fileout is : exampleout T - d.save() #save currently viewed spectrum to output file.
Parameters:
keep does the same thing as save in the default case; it takes no arguments.
lsoutfile - List the scan numbers in the specified output file.
Example:
- d.fileout('exampleout')
New file exampleout is created
- d.save()
- d.lsoutfile()
159
writetofile - Write ASCII columned data of x and y to a disk file.
Example:
- d.writetofile('final.spc'); #default is to write currently viewed spectrum.
The columns that are written out are: 1) the x-axis in the currently displayed units and 2)-n) the data arrays for each polarization/receptor.
Parameters:
Imaging in dish is done with the imager tool in AIPS++. Currently, there is a single function makeim in dish to achieve this.
makeim - Make a spectral line data cube.
Example:
- d.makeim('final.spc'); #default is to write currently viewed
This function selects the appropriate data from the MeasurementSet, regrids the data and constructs the image. The data are converted into the coordinate system of the image and then added to the image using a gridding function (BOX, SF, or PB). It also constructs a coverage image which is simply the gridded weights; this is used to normalize the data for the sampling density - the final output image is the image name with a "_corr" attached (imname_corr).
Parameters:
The image tool function moments offers traditional moment analysis (weighted sums of profiles) of images. It offers a wide range of moments and a wide range of methods with which to calculate them. The word `moment' is used loosely here. It refers to collapsing an axis (the moment axis) to one pixel and setting the value of that pixel (for all of the other non-collapsed axes) to something computed from the data values along the moment axis. For example, take an RA-DEC-Velocity cube, collapse the velocity axis by computing the mean intensity at each RA-DEC pixel. This function offers many different moments and a variety of interactive and automatic methods to compute them. A quick summay of the available moments:
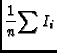
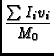
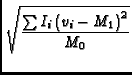
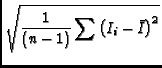
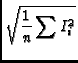
- im:=image('g29im') #g29im is the data cube
- im.moments(moments=[-1,0]) #get the average and integrated moments
Moment axis type is Frequency
***********************************************************************
You have selected the following methods
The basic method
Created /home/despina2/scratch/jmcmulli/g29im.average
Created /home/despina2/scratch/jmcmulli/g29im.integrated
Begin computation of moments
Finished image::moments
0.48 real 0.08 user 0.02 system
T
- im2:=image('g29im_integrated')
- im2.view()
- im.summary();
- r:=drm.box(blc='1 1 1 5', trc='20 20 1 100') # create a region
- #using the channels 5 to 100; the first 2 coordinates are ra, dec,
- #the 3rd is stokes, and the 4th is the channel axis.
- im.moments(moments=0,region=r,outfile='g29im_lowvel') #create the
- #integrated intensity over channels 5-100
You can also use the GUI interface via:
- im:=image('g29im') #g29im is the data cube
- im.momentsgui(); #use GUI to derive moments
See the User Reference Manual on the moments function for the most details moments and moment methods.
Source fitting is achieved through the imagefitter tool. From the example above:
- imfit:=imagefitter('g29im_integrated')
- #Use the GUI to select a region (double click to derive source characteristics,
- #statistics, residuals, etc. The 'show' button will display the fit value.
Manipulation of images is done with the imagecalc tool. With this you can construct an image from expressions involving other images. For example,
im1 := imagecalc(outfile='x3', pixels='sin(x1) + min(x2) ')
where the image disk files x1 and x2 must pre-exist and the result is the image disk file called x3.
You can also construct a read-only image from expressions involving other images. The output image is never created, it is just evaluated each time you use it.
im2 := imagecalc(pixels='pi() - min(x2) + sqrt(x1)')
where the image disk files x1 and x2 must pre-exist. Note that by simply excluding the output file name (argment outfile) you have created the read-only image.
With the image calc function, you can change the pixel values of an image by replacing them with the result of the expression:
im1 := image('x2')
im2 := im1.calc('abs(x2) / min(x2)')
where the image disk file x2 must pre-exist. Because the scalar expression is evaluated first, the images in the expression can be the same as the image which is being overwritten.
Note that for image file names with special characters in them (like for example), you should (double) escape those characters or put the file name in ouble quotes. E.g.
- im1 := imagecalc(pixels='test\\-im') # Note double escape - im2 := imagecalc(pixels='"test-im"')
When using imagecalc, you can use either image disk file names, or an associated image tool name in the expression. For example,
im1 := image('im1')
im2a := imagecalc('im2a', '2*im1') # Use disk file names
im2b := imagecalc('im2b', '2*$im1') # Use tool name
For information on image manipulation (concatenation, addition, etc), see the Getting Results on Image Analysis.
To exit dish, type 'exit' at the Glish command line. If you have enabled the "Save state when done" option, the state of dish will be preserved for your next session (it will be loaded automatically). You can also manually save the state of dish using the File menu options of the main dish GUI.
![]()
aver Average a group of scans,subscans.
base Perform a polynomial baseline fit to a scan.
baseline Perform a polynomial baseline fit to spectrum in globalscan1
bdrop Set value for dropping beginning channels of a spectrum in display
boxcar Perform a boxcar smoothing
bshape Show the baseline fit of order nfit to globalscan1
calib Apply calibration to a scan (GBT only currently).
clearrm Clear all variables from results manager, etc.
clip Clip/Edit data (set flags)
done Exits and destroys the current Dish tool.
edrop Set value for dropping end channels of a spectrum in display
filein Sets the scan group that will be manipulated.
fileout Sets the scan group that can be written to.
files Prints the currently set values for filein/out.
find Do a grep on a string to find glish variables.
galactic Provide the coordinates in the Galactic reference frame.
gauss Fit gaussian profiles.
getc Retrieve calibrated data (from the CORRECTED\_DATA column)
getr Retrieve raw data (from the FLOAT\_DATA column)
getscan Retrieve a scan (SDRecord) from a scan group.
getvf Return the velocity and frequency for a specified channel.
getvfarray Return the velocities and frequences for the currently viewed
spectrum.
gms Print MeasurementSet summary to screen.
gui Toggles mapping of DISH main gui (results manager).
hanning Perform a hanning smooth
header Print formatted header information to the command line window
help Provide simple help on dish functions
history Add a string or vector of strings to the history of a current SDRecord
imcal Calibrate an imaging/mapping procedure
import Import GBT data into an AIPS++ MeasurementSet
keep Saves the current globalscan1 SDRecord to the designated fileout.
listscans List the scan numbers from the active scan group.
logcommand Writes command to scripter.
lsoutfile Lists scans in the fileout scangroup
makeim Make an image from scans in a MeasurementSet.
message Writes a command to the DISH GUI message entry.
mscal Calibrate a MeasurementSet.
mult Scales (multiplies) a scan by a value.
nfit Set the order of the polynomial for baseline subtraction.
nogui Unmaps frame to DISH main GUI.
nregion Set the channel ranges for baseline subtraction.
open Load a scan group from disk into DISH and perform a fil ein on it.
page Clears the DISH plotter display.
peak Obtain the peak amplitude, half-width and center in the displayed
spectrum
plotc Plot calibrated data (from the CORRECTED\_DATA column)
plotr Plot raw data (from the FLOAT\_DATA column)
plotscan Plot an SDRecord.
plotxy Plot two vectors in the DISH plotter.
qscan Query characteristics of scan.
reference See TPcal
restore Retrieve the contents of the uni Record from disk.
restorestate Restore the state of DISH's GUIs.
rmadd Add an SDRecord to the results manager.
rms Display statistics on baseline regions.
save Save an SDRecord to file.
savestate Save the current state of DISH.
saxis Set the units of the plotters displayed x axis
scale Multiply the array in an SDRecord by a value.
scanadd Add scan1 and scan2.
scanbias Adds a bias level to a scan.
scandiv Divide scan 1 by scan 2.
scanmult Multiply scan 1 by scan 2.
scanscale Scale a scan by a value.
scansrr Performs a signal-reference/reference.
scansub Subtract scan2 from scan1.
select Select a subset of a scangroup.
setregion Set the channel range for use in baseline subtraction
show Display the globalscan1 SDRecord
show1 Display one polarization of the globalscan1 SDRecord
signal See TPcal
smooth Smooth (hanning, boxcar or gaussian) an SDRecord.
stat Generate statistics over a line region.
statefile Changes file for state saving.
stats Generate statistics over a region.
store Store the contents of the uni Record on disk.
summary Print out a more detailed summary of scans from a scan group.
upr Print the current value of the specified variable in the uni-record.
utable Print channel and data columns to command window
writetofile Write X,Y axes to an ascii file on disk.
In the following, the > designates a UNIX prompt, and a '-' designates the glish prompt.
This section details calibration of data taken in the following modes:
> source /home/aips++/gbt/aipsinit.sh #load environment
> dish #start AIPS++ DISH tool
...
- d.open('exampleSP')
T
- d.files() #show active files
Current filein is : exampleSP
Current fileout is : not set
T
- d.gms() #provide brief summary
Scan Object Proctype SWState SWtchsig Procseqn Procsize
35 S8 Track FSWITCH FSW01 1 1
36 S8 Track FSWITCH FSW12 1 1
37 S8 Track FSWITCH FSW12 1 1
38 S8 OffOn PSWITCHOFF TPWCALSP 1 2
40 S8 OnOff PSWITCHON TPWCALSP 1 2
42 S8 Track NONE TPWCALSP 1 1
T
- #Note: This only lists the first scan in a procedural sequence of scans
- d.listscans() #shows all scans present
[35 36 37 38 39 40 41 42]
- d.plotr(35,1) #plot the raw(observed) data
#for the first phase of scan
#35. Since no integration
#value is specified, it averages
#all phase 1 data (in this case
#this is all data with CAL ON.
-
- d.calib(35) #this will calibrate scan 35
#according to the way the
#the data were taken (i.e., the
#observing procedure
- d.plotc(35) #plot the calibrated data for
#scan 35. Since no integration
#was specified, all data is
#averaged.
- d.plotc(35,1) #plots the first integration of
#the calibrated data for 35.
- d.plotc(35,3,2) #plots the third integration of
#the second polarization for
#scan 35.
- d.plotc(scan=35,int=3,pol=2) #same using the explicit names
#for the arguments.
-
- #HINT: if you are uncertain as to the arguments for a function, simply type
- # the function name.
- d.plotc
function (val scan, val int = F, val pol = F) {
(ok := public.plotscan(public.getc(scan, int, pol)));
return T}
- #NOTE: if you are unhappy with the calibration, you can simply reapply it
- # using different attributes
- d.calib(35,baseline=T,range=[300:600,800:1000]);
#Apply calibration but also
#baseline the data using the
#specified range (defaults
#to first order but this can
#also be specified.
- #This will over-write the calibration data with the new calibration data.
- d.plotc(35) #re-examine the data
This section details calibration of data taken in the following modes:
> source /home/aips++/gbt/aipsinit.sh #load environment
> dish #start AIPS++ DISH tool
...
- d.open('exampleSP')
T
- d.files() #show active files
Current filein is : exampleSP
Current fileout is : not set
T
- d.gms() #provide brief summary
Scan Object Proctype SWState SWtchsig Procseqn Procsize
35 S8 Track FSWITCH FSW01 1 1
36 S8 Track FSWITCH FSW12 1 1
37 S8 Track FSWITCH FSW12 1 1
38 S8 OffOn PSWITCHOFF TPWCALSP 1 2
40 S8 OnOff PSWITCHON TPWCALSP 1 2
42 S8 Track NONE TPWCALSP 1 1
T
- #Note: This only lists the first scan in a procedural sequence of scans
- d.listscans() #shows all scans present
[35 36 37 38 39 40 41 42]
- d.plotr(42,1) #plot the raw(observed) data
#for the first phase of scan
#42. Since no integration
#value is specified, it averages
#all phase 1 data (in this case
#this is all data with CAL ON.
-
- d.calib(42) #this will calibrate scan 42
#according to the way the
#the data were taken (i.e., the
#observing procedure
- d.plotc(42) #plot the calibrated data for
#scan 42. Since no integration
#was specified, all data is
#averaged.
- d.plotc(42,1) #plots the first integration of
#the calibrated data for 42.
- d.plotc(42,3,2) #plots the third integration of
#the second polarization for
#scan 42.
- d.plotc(scan=42,int=3,pol=2) #same using the explicit names
#for the arguments.
-
- #HINT: if you are uncertain as to the arguments for a function, simply type
- # the function name.
- d.plotc
function (val scan, val int = F, val pol = F) {
(ok := public.plotscan(public.getc(scan, int, pol)));
return T}
- #NOTE: if you are unhappy with the calibration, you can simply reapply it
- # using different attributes
- d.calib(42,baseline=T,range=[300:600,800:1000]);
#Apply calibration but also
#baseline the data using the
#specified range (defaults
#to first order but this can
#also be specified.
- #This will over-write the calibration data with the new calibration data.
- d.plotc(42) #re-examine the data
- #NOTE: You can also individually calibrate each scan in the sequence with
- # 'd.TPcal(scan)'. There are numerous {\bf dish} functions for
- # manipulating SDRecords (e.g., {\it scanscale (multiplies a scan
- # by a factor, scandiv (divides two scans), etc.
> source /home/aips++/gbt/aipsinit.sh #load environment
> dish #start AIPS++ DISH tool
...
- d.open('exampleSP')
T
- d.files() #show active files
Current filein is : exampleSP
Current fileout is : not set
T
- d.gms() #provide brief summary
Scan Object Proctype SWState SWtchsig Procseqn Procsize
35 S8 Track FSWITCH FSW01 1 1
36 S8 Track FSWITCH FSW12 1 1
37 S8 Track FSWITCH FSW12 1 1
38 S8 OffOn PSWITCHOFF TPWCALSP 1 2
40 S8 OnOff PSWITCHON TPWCALSP 1 2
42 S8 Track NONE TPWCALSP 1 1
T
- #Note: This only lists the first scan in a procedural sequence of scans
- d.listscans() #shows all scans present
[35 36 37 38 39 40 41 42]
- d.plotr(39,1) #plot the raw(observed) data
#for the first phase of scan
#38. Since no integration
#value is specified, it averages
#all phase 1 data (in this case
#this is all data with CAL ON.
-
- d.calib(39) #this will calibrate scan 38/39
#(since both compose the
#the OffOn procedure),
#according to the way the
#the data were taken (i.e., the
#observing procedure
- d.plotc(39) #plot the calibrated data for
#scan 39. Since no integration
#was specified, all data is
#averaged.
#The (sig-ref)/ref data is
#only stored in the 'On' scans
#data record; the 'Off' scan
#contains only the calibrated
#TPwCal data.
- d.plotc(39,1) #plots the first integration of
#the calibrated data for 39.
- d.plotc(39,3,2) #plots the third integration of
#the second polarization for
#scan 39.
- d.plotc(scan=39,int=3,pol=2) #same using the explicit names
#for the arguments.
-
- #HINT: if you are uncertain as to the arguments for a function, simply type
- # the function name.
- d.plotc
function (val scan, val int = F, val pol = F) {
(ok := public.plotscan(public.getc(scan, int, pol)));
return T}
- #NOTE: if you are unhappy with the calibration, you can simply reapply it
- # using different attributes
- d.calib(38,baseline=T,range=[300:600,800:1000]);
#Apply calibration but also
#baseline the data using the
#specified range (defaults
#to first order but this can
#also be specified.
- #This will over-write the calibration data with the new calibration data.
- d.plotc(38) #re-examine the data
> source /home/aips++/gbt/aipsinit.sh #load environment
> dish #start AIPS++ DISH
tool
...
- d.open('exampleSP')
T
- d.files() #show active files
Current filein is : exampleSP
Current fileout is : not set
T
- d.gms() #provide brief summary
Scan Object Proctype SWState SWtchsig Procseqn Procsize
35 S8 Track FSWITCH FSW01 1 1
36 S8 Track FSWITCH FSW12 1 1
37 S8 Track FSWITCH FSW12 1 1
38 S8 OffOn PSWITCHOFF TPWCALSP 1 2
40 S8 OnOff PSWITCHON TPWCALSP 1 2
42 S8 Track NONE TPWCALSP 1 1
T
- #Note: This only lists the first scan in a procedural sequence of scans
- d.listscans() #shows all scans present
[35 36 37 38 39 40 41 42]
- d.plotr(39,1) #plot the raw(observed) data
#for the first phase of scan
#38. Since no integration
#value is specified, it averages
#all phase 1 data (in this case
#this is all data with CAL ON.
-
- d.calib(39) #this will calibrate scan 38/39
#(since both compose the
#the OffOn procedure),
#according to the way the
#the data were taken (i.e., the
#observing procedure
- d.plotc(39) #plot the calibrated data for
#scan 39. Since no integration
#was specified, all data is
#averaged.
#The (sig-ref)/ref data is
#only stored in the 'On' scans
#data record; the 'Off' scan
#contains only the calibrated
#TPwCal data.
- d.plotc(39,1) #plots the first integration of
#the calibrated data for 39.
- d.plotc(39,3,2) #plots the third integration of
#the second polarization for
#scan 39.
- d.plotc(scan=39,int=3,pol=2) #same using the explicit names
#for the arguments.
- #Now, if you wanted to use a different reference scan for this on, you could
- #do the following:
- d.TPcal(41) #calibrate 'Off' scan 41.
- d.SRcal(39,41) #perform (sig-ref)/ref with
#signal=39, reference=41
- d.SRcal(39,[38,41]); #perform (sig-ref)/ref with
#signal scan=39, reference=
#the average of 38 and 41.
#in general:
-d.SRcal([vector of signal scans],
[vector of reference scans]) #all reference scans will be
#averaged and subtracted from
#each signal scan.
> source /home/aips++/gbt/aipsinit.sh #load environment
> dish #start AIPS++ DISH tool
...
- d.open('exampleSP')
T
- d.files() #show active files
Current filein is : exampleSP
Current fileout is : not set
T
- d.gms() #provide brief summary
Scan Object Proctype SWState SWtchsig Procseqn Procsize
303 S6 Track FSWITCH FSW01 1 1
304 G250p8 RALongMap FSWITCH FSW01 1 41
T
- #Note: This only lists the first scan in a procedural sequence of scans
- d.listscans() #shows all scans present
[303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321
322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340
341 342 343 344]
- d.plotr(304,1) #plot the raw(observed) data
#for the first phase of scan
#303. Since no integration
#value is specified, it averages
#all phase 1 data (in this case
#this is all data with CAL ON.
-
- d.calib(304) #this will calibrate scan 304
#according to the way the
#the data were taken (i.e., the
#observing procedure
- d.plotc(303) #plot the calibrated data for
#scan 303. Since no integration
#was specified, all data is
#averaged.
- d.plotc(303,1) #plots the first integration of
#the calibrated data for 303.
- d.plotc(303,3,2) #plots the third integration of
#the second polarization for
#scan 303.
- d.plotc(scan=303,int=3,pol=2); #same using the explicit names
#for the arguments.
-
- #HINT: if you are uncertain as to the arguments for a function, simply type
- # the function name.
- d.plotc
function (val scan, val int = F, val pol = F) {
(ok := public.plotscan(public.getc(scan, int, pol)));
return T}
- #NOTE: if you are unhappy with the calibration, you can simply reapply it
- # using different attributes
- d.calib(303,baseline=T,range=[300:600,800:1000]);
#Apply calibration but also
#baseline the data using the
#specified range (defaults
#to first order but this can
#also be specified.
- #This will over-write the calibration data with the new calibration data.
- d.plotc(303) #re-examine the data
- #NOTE:In addition, if the SIG/REF signals are mixed (there is a temporary
- # problem with the SPECTRAL PROCESSOR which causes some fraction of the
- # scans to be flipped), you can reverse the sense of SIG and REF by
- # doing:
- d.FScal(304,flipsr=T) #flips what is SIG and REF
-
- #NOTE:You can write a procedure to interactively view and flip your data.
-
- sns:=seq(304,344) #make a vector from 304-344
- for (i in sns) { #loop over these scans
+ d.plotc(i) #plot scan
+ ans:=readline() #pause and prompt user for input
+ if (ans=='y') { #if 'y' typed, then
+ d.FScal(i,flipsr=T) # flip SIG and REF sense
+ }
+ }
-
- d.makeim(304,600,900,imname='ralong') #Make an image. This needs the
# first scan in the map
# the beginning (600) and end
# (900) channels to include.
# step determines the
# averaging over channel
# (none here) and imname
# specifies the output image
# NOTE: This actually creates three images on disk: 1) the original image,
# a weights image, and an image corrected (divided) by the weights.
# These are imname, imname\_weight and imname\_corr.
- im:=image('ralong\_corr') #Make an image tool - lots of
#functions here!
- im.view() #View the image
Currently, the DCR tool is in a separate utility and must be included.
include 'gbtcalutils.g';This creates a gc tool which four associated functions:
See the Getting Results chapter on Continuum Single Dish Imaging for more details: docs/gettingresults/grvol3/node4.html.
- gc.setdata('cassaDCR');
successful readonly open of default-locked table cassaDCR/NRAO\_GBT\_GLISH: 21 col
umns, 120 rows
successful readonly open of default-locked table cassaDCR/NRAO\_GBT\_IF: 22 column
s, 240 rows
successful read/write open of default-locked table cassaDCR/FEED: 12 columns, 2
rows
successful read/write open of default-locked table cassaDCR/POLARIZATION: 4 colu
mns, 2 rows
successful read/write open of default-locked table cassaDCR/SPECTRAL\_WINDOW: 14
columns, 1 rows
successful readonly open of default-locked table cassaDCR/POINTING: 15 columns,
48000 rows
T
- gc.contcal()
successful read/write open of default-locked table cassaDCR: 28 columns, 192000
rows
T
- gc.makeimage()
successful readonly open of default-locked table cassaDCR: 28 columns, 192000 ro
ws
Starting server imager
Server started: /export/home/guthwine/aips++/linux\_gnu/bin/imager (AIPS++ versio
n: 1.7 (build #405))
Opening MeasurementSet cassaDCR
defaultcatalog (dc) ready
Starting imager::setdata
Selecting data
Performing selection on MeasurementSet
Selecting on field and spectral window ids
By selection 192000 rows are reduced to 48000
Finished imager::setdata
0.96 real 0.69 user 0.19 system
successful readonly open of default-locked table cassaDCR/NRAO\_GBT\_GLISH: 21 col
umns, 120 rows
Cell size is 0.289495253arcmin
Map size is 117.705376 by 59.9669179
Grid sizes are 406 208
Defining image properties
nx = 406 is not composite; nx = 432 or 400 will be more efficient
ny = 208 is not composite; ny = 216 or 200 will be more efficient
Setting processing options
Starting imager::weight
Weighting MS: IMAGING\_WEIGHT column will be changed
Natural weighting
Sum of weights = 48000
Finished imager::weight
32.6 real 23.58 user 6.11 system
Starting imager::makeimage
Calculating image (without full skyequation)
Making single dish image from corrected data
Image is : scanimage
Frequency = 8.9 GHz, synthesized continuum bandwidth = 0.08 GHz
Image polarization = Stokes I
Frequency = 8.9 GHz, synthesized continuum bandwidth = 0.08 GHz
Image polarization = Stokes I
Performing Single Dish gridding with convolution function SF
Gridding will use specified common tangent point:
23:21:00.00 +58.32.00.00 B1950
Using prolate spheriodal wave function as the convolution function
Finished imager::makeimage
10.25 real 6.77 user 0.49 system
Starting imager::makeimage
Calculating image (without full skyequation)
Making single dish coverage function
Image is : scanimage\_weight
Frequency = 8.9 GHz, synthesized continuum bandwidth = 0.08 GHz
Image polarization = Stokes I
Frequency = 8.9 GHz, synthesized continuum bandwidth = 0.08 GHz
Image polarization = Stokes I
Using prolate spheriodal wave function as the convolution function
Finished imager::makeimage
8.12 real 6.58 user 0.52 system
Successfully closed empty server: imager
T
- gc.covercorr()
Starting server app\_image
Server started: /export/home/guthwine/aips++/linux\_gnu/bin/app\_image (AIPS++ ver
sion: 1.7 (build #405))
Selected bounding box :
[1, 1, 1, 1] to [406, 208, 1, 1] (23:28:23.897, +58.01.04.977, I, 8.900000e
+09Hz to 23:13:25.534, +59.00.59.643, I, 8.900000e+09Hz)
Creating new statistics storage lattice of shape [9]
Number points = 8.444800e+04 Sum = 8.444801e+04
Mean = 1.000000e+00
Variance = 1.026810e+00 Std dev = 1.013316e+00
Rms = 1.423657e+00
Minimum value 0.000000e+00 at [1, 1, 1, 1] (23:28:23.897, +58.01.04.977, I, 8.
900000e+09Hz)
Maximum value 4.499732e+00 at [105, 182, 1, 1] (23:24:41.961, +58.54.05.639, I
, 8.900000e+09Hz)
Creating image `scanimage\_corr' of shape [406, 208, 1, 1]
Created mask `mask0'
T
- #Scale images to Jy (e.g., 1.76 K/Jy)
- im:=imagecalc(outfile='scanimage\_tmb',pixels='(scanimage\_corr)/1.76');
- im.view(); #note the higher amplitudes
Because AIPS++ is an actively developing software system, there are several versions available. These are called release, stable, weekly and daily. In Green Bank we maintain an additional version called gbt, which is just a link to the best version for reducing GBT data.
To start stable dish, issue the following commands (from the bash shell):
. /home/aips++/gbt/aipsinit.sh dish
or for (t)csh:
. /home/aips++/gbt/aipsinit.sh dish
To run one of the other versions, simply replace the word gbt in the path with the name of the version required. It is occassionally necessary to run weekly or daily to incorporate the latest bug fixes or enhancements.
You can run AIPS++ on any Linux system in Green Bank, but if the operation seems sluggish to you here are some hints on how to speed things up:
user.aipsdir: /tmp/username\_aips++
where username is your user name, e.g.
user.aipsdir: /tmp/jbraatz\_aips++
toolmanager.fullgui: F
toolmanager.gui.auto: F
The toolmanager GUI can then be restarted at a later time by:
- tm.gui()
ssh despina cd /export/home/despina3/scratch mkdir my\_dir cd my\_dir aips++
Keep in mind that AIPS++ has to build its cache file structure the first time you start it with a new user.aipsdir, and each time you start a new version. So judge its startup speed on the second time you start it up, not the first. On etamin it should take about 14 seconds to start AIPS++ with the toolmanager GUI, and 7 seconds without it.
Make yourself a data directory starting at the following:
despina: /export/home/despina3/scratch thalassa: /export/home/thalassa2/scratch naiad: /export/home/naiad3/scratch etamin: /export/home/etamin/scratch general use: /home/aips++\_data (slow access from here)
If you find that error messages, code, and data listings go flying across your terminal window, and you'd like to page them instead (as with the more or less programs) then it may be that the PAGER environment variable is not set properly on your system. This seems to be the case for many of our systems in GB. You can remedy it by setting PAGER to "less" in your UNIX startup scripts. Contact Jim if you need help doing so. It makes glish nicer!
This section shows an example data reduction session using dish in a UniPOPS fashion.
% ssh etamin
% cd /home/etamin/scratch
% mkdir myname
% cd myname
% . /home/aips++/gbt/aipsinit.sh
% aips++ -l uni2.g
- d.import('/home/gbtdata/tigerTeam\_02','tiger.ms',startscan=20,stopscan=33)
- d.lscans()
[20 21 22 23 24 25 26 27 28 29 30 31 32 33]
- d.reference(20)
- d.signal(21)
- d.temp()
- d.show()
- d.nfit(5)
# Use the plotter to set the x-axis units to pixels (channel number)
- d.nregion(0,3600,4100,8200)
- d.bshape() # to show the fit
- d.baseline() # to subtract the baseline
- d.show()
- d.uscale(2.3)
- d.setYUnit('Jy')
- d.show()
- d.bdrop(2500)
- d.edrop(2500)
- d.show()
- d.sclear()
- d.getc(26)
- d.show()
- d.stats(bchan=1000,echan=2000)
- d.accum()
- d.getc(28)
- d.show()
- d.accum()
- d.stats(bchan=1000,echan=2000)
- d.ave()
- d.show()
- d.stats(bchan=1000,echan=2000)
The tigerTeam_02 data in the examples used throughout this document were taken with the GBT on 11 October 2001. The L-band receiver (1.15-1.73 GHz) was used with the intent to observe the HI profiles of an assortment of galaxies. The backend was the GBT spectrometer. The ``OffOn'' procedure in GO was used to drive the position-switched observations. Signal and reference data are recorded in separate, adjacent scans of equal duration.
The first step in processing data is to identify the input data file. The technique for doing so differs depending on if the data are in the raw GBT FITS format, or have been previously filled into the AIPS++ MeasurementSet format. To import data from FITS files, use the import command:
- d.import('/home/gbtdata/tigerTeam\_02','tiger.ms',startscan=20,stopscan=33)
This command creates a MeasurementSet called tiger.ms. If the desired
MeasurementSet already exists on disk, use the dopen command to load the data:
- d.open('/home/aips++/data/demo/dishdemo/tiger.ms')
These data sets are specific to the Green Bank installation. For other sites, consult the AIPS++ system administrator to see how to access this data.
List the scans available in your new data set:
- d.lscans() [20 21 22 23 24 25 26 27 28 29 30 31 32 33]
Calibrate a Sig/Ref pair as follows:
- d.reference(20) - d.signal(21) - d.temp() - d.show()
You can also use the core dish functions to calibrate scans.
- d.calib(20) or - d.TPcal(20) - d.TPcal(21) - d.SRcal(21,20) - d.plotc(21)
To calibrate and retrieve a frequency switched scan, only a single get is required:
# This example does not work with the sample data provided, but shows the # sequence required for freq switched data. - d.get(2005) - d.show()
The red and green plots represent the two polarizations. These are currently handled together (use the getpol function to retrieve a single polarization. Currently you can show an individual polarization with the show1 function, and you can average to two polarizations using the avgFeeds function. The spectrum is initially shown with GHz units for the x-axis. These units can be changed using the button on the plotter labeled ``GHz''. Regions of the plot may be zoomed using the middle mouse button, and unzoomed using the unzoom button on the plotter.
To fit a baseline to the data, you must specify a range of the spectrum to use for the fit and the order of the polynomial. In this example we will describe the fit region using channel numbers, so first plot the spectrum against channel number (select the option labeled pix on the plotter x-axis selector button). Then, similar to UniPOPS, use the following procedure:
- d.nfit(5) - d.nregion(0,3600,4100,8200) - d.bshape() # to show the fit - d.baseline() # to subtract the baseline - d.show() # to show the baseline-subtracted data
The UNI-jr function setregion() can be used in place of the nregion function to enter the baseline region interactively using the mouse cursor.
If a calibration scaling factor is known, it can be applied:
- d.uscale(2.3)
- d.setYUnit('Jy')
- d.show()
The UNIPOPS-familiar bdrop and edrop parameters may be utilized in the plots:
- d.bdrop(2500) - d.edrop(2500) - d.show()
To recall the value of a UNIPOPS-type adverb, simply call the function with no value, as follows:
- d.bdrop() 2500 - d.nfit() 5
To fit a gaussian function to the data, use the ``G fit'' button on the plotter. Prior to fitting, an initial guess to the line shape must be given. This can be done automatically by pressing the green ``Estimate'' button, or interactively by pressing the gray ``Inter'' button under the ``Estimates'' title. Choose the latter for the sample data and follow the directions on the message line in the Profile Fitter GUI. Then press the green ``Fit profile'' button the do the actual fitting.
UNI-jr uses the UNIPOPs model for averaging scans. Data is stored away to the accumulator, and can then be averaged. First clear the accumulator:
- d.sclear()
then retrieve scans using (for example) get, getc, signal/reference/temp, getfs, or some other calibration and retrieval command, and accumulate them:
- d.getc(26) - d.accum() - d.getc(28) - d.accum()
and finally average and show the result:
- d.ave() - d.show()
The stats function will return statistics over a given channel range:
- d.stats(bchan=500,echan=1500)
The equivalent to the UNIPOPS moment command can be run as follows:
- d.bmoment(3900) - d.emoment(4100) - d.moment()
This section lists the commands currently available in the UNI-jr package. Each function's parameters are listed with it. If the parameter does not have an assignment (e.g. the scan parameter in the signal function) then it must be supplied when the function is called. If the parameter does have an assignment, then the assigned value shown here is the default, but it can be overridden by specifying a new value when the function is called.
The functions can either be called with parameters listed in order and no parameter name specified, or they can be called with parameter names specified and in any order. The following two calls are equivalent:
d.stats(F,2,1000,2000)
d.stats(bchan=1000,echan=2000,feed=2)
Loading data:
------------
d.import(projdir,outms=F,outmsdir=F,startscan=F,stopscan=F,backend=F)
: Import (fill) data from GBT FITS files
d.open(filename) : Open an already filled data set
d.files() : Show input and output file settings
d.summary() : List some scan info (source, time)
d.ls() : List files in directory
Retrieving and Calibrating Scans:
---------------------------------
d.lscans() : List available scans
d.dgetscan(scan,nphase) : Return an individual phase
d.getir(scan,phase) : Get an individual phase to memory
d.get(scan) : General purpose scan retrieval
d.signal(scan,tcal\_in=unset,pctTsys=0.01,verbose=F)
: Retrieve a sig scan
d.reference(scan,tcal\_in=unset,pctTsys=0.01,verbose=F)
: Retrieve a ref scan
d.temp() : Perform (s-r)/r for pos switch data
d.getc(scan) : Retrieve \& calibrate pos switch pair
d.getfs(scan,tcal\_in=unset,flipsigref=F,pctTsys=0.001,verbose=T)
: Retrieve freq switch scan
d.getfsint(scan,intNo=1,tcal\_in=unset,flipsigref=F,pctTsys=0.01,verbose=F)
: Retrieve freq switch integration
Averaging Scans:
----------------
d.accum() : Add scan to accumulator
d.ave() : Average scans in accumulator
d.sclear() : Clear accumulator
d.avgFeeds() : Average all feeds
Plotting Scans:
----------------
d.page() : Clear the plotter
d.show() : Display scan in memory (globalscan1)
d.show1(ifeed) : Display 1 feed of scan in memory
d.showref() : Display ref scan
d.setYUnit(unit) : Set Y axis unit for plotting
d.saxis(strval) : Change units on X axis
d.bdrop(value=T) : Set bdrop parameter
d.edrop(value=T) : Set edrop parameter
Baseline Fit:
-------------
d.nfit(value=T) : Set order of polynomial to fit
d.nregion(...) : Set baseline region
d.setregion() : Set baseline region with mouse
d.baseline() : Subtract polynomial of order nfit
d.dcbase() : Subtract polynomial of order 0
d.bshape() : Show baseline fit of order nfit
Smoothing and Scaling data:
---------------------------
d.uscale(factor) : Scale the scan in memory
d.bias(factor) : Add a bias to scan in memory
d.hanning() : Hanning smooth scan in memory
d.boxcar(smooth\_width) : Boxcar smooth scan in memory
d.chngres(smooth\_width) : Change resolution of scan in memory
The Stack:
----------
d.addstack(beg,end=beg,inc=1) : Add elements to the stack
d.empty() : Empty the stack
d.delete(value) : Delete one component from the stack
d.tellstack() : Show the stack
Analysis:
---------
d.stats(quiet=F,feed=0,bchan=0,echan=0) : Calculate stats over specified range
d.rms(printFlag=F) : Calc stats over the baseline region
d.bmoment(value=T) : Set bmoment parameter
d.emoment(value=T) : Set emoment parameter
d.moment(quiet=F) : Calculate moments of scan in memory
d.peak(quiet=F) : Calculate location and val of peak
d.galactic(quiet=F) : Calculate galactic coords from header
Scan Arithmetic:
----------------
d.push() : push a scan for processing
d.minus() : Perform sig = sig-ref
d.plus() : Perform sig = sig+ref
d.multiply() : Perform sig = sig*ref
d.divide() : Perform sig = sig/ref
Saving Data:
------------
d.fileout(outfile=F) : Set output file
d.keep() : Save a scan
d.lsoutfile() : List scans in keep file
Utility:
--------
d.lscans() : List available scans
d.utable() : Show data values from scan in memory
d.header() : Show header values for scan in memory
d.getvf(chan) : Show velocity, freq for given channel
d.getvfarray() : Get array or velocities, frequencies
d.uniput(name,value) : Store a value in memory slot
d.uniget(name) : Retrieve a value from memory slot
d.copy(fromhere,tohere) : Copy memory locations
d.upr(...) : Show internal variable values
d.store() : Store any state values (nregion, ...)
d.restore() : recall stored values
UNIPOPS {\bf dish}
------- ------
get 55 d.get(55)
page d.page()
show d.show()
accum d.accum()
ave d.ave()
sclear d.sclear()
nfit = 3 d.nfit(3)
nregion = 1,128,380,512 d.nregion(1,128,380,512)
baseline d.baseline()
dcbase d.dcbase()
bshape d.bshape()
on 55 d.signal(55)
off 56 d.reference(56)
temp d.temp()
factor=3.5; scale d.uscale(3.5)
factor=1.2; bias d.bias(1.2)
addstack(600,620,1) d.addstack(600,620,1)
addstack(631) d.addstack(631)
empty d.empty()
delete(609) d.delete(609)
tell stack d.tellstack()
table d.utable()
header d.header()
bmoment = 128 d.bmoment(128)
emoment = 512 d.emoment(512)
moment d.moment()
minus d.minus()
plus d.plus()
multiply d.multiply()
divide d.divide()
peak() d.peak()
chngfile d.chngfile()
keep() d.save()
hanning d.hanning()
nbox=5; boxcar d.boxcar(5)
newres=4.4; chngres d.chngres(4.4)
The standards data set used for this example is:
/home/gbtdata/standards_apr27
and represents only Spectral Processor data.
The following details the relevant observing modes and scan numbers:
| Scan | Procedure | Source | Switch State | Switch Signal | BE |
| 5 | OffOn | S171 | PSWITCHOFF | TPWCAL | SP |
| 6 | OffOn | S171 | PSWITCHON | TPWCAL | SP |
| 7 | OffOn | S171 | PSWITCHOFF | TPWCAL | SP |
| 8 | OffOn | S171 | PSWITCHON | TPWCAL | SP |
| 9 | Track | S171 | FSWITCH | FSW01 | SP |
| 15 | RALongMap | OrionA | FSWITCH | FSW01 | SP |
For the GBT users, updates to the AIPS++ releases are supported through a binary inhale. This option is only available for Linux users (Solaris is not supported). If the user has an AIPS++ release installed (from a CD or ISO image), he may 'update' this release to a newer version of the code through the following procedure:
1. On a linux system (binary inhale is not available for solaris), install
aips++ v1.7 from a release CD. Install the bare-minimum release. If
you want the up-to-date documentation, it can be retrieved below.
2. Change directory to the aips++ root directory (the directory
you specified during the INSTALL procedure).
3. Make a symbolic link called linux_gnu that points to the linux
directory (which contains the bulk of the release installation):
% ln -s linux linux_gnu
4. Edit the .aipsrc file as follows:
a. Comment out all lines referring to machines in Socorro (tarzan,
cluster)
b. Add the following line (replacing <username> with your username)
account.manager: <username>
5. Source the aips++ initialization script:
% source ./aipsinit.csh
6. Add the current directory to your PATH:
% setenv PATH .:$PATH
7. Initiate the inhale:
a. To retrieve only the code and binaries:
% inhale binary
b. To retrieve the data repository and documentation, as
well as the code and binaries (be sure 'cvsup'
is available on your machine):
% inhale -cvsup <x> binary
(where <x> is data, docs, or all, depending upon what you want)
This will take up to several hours while a snapshot of the latest
aips++ stable installation is retrieved (from Socorro), unpacked, and
assembled.
Subsequent binary inhales require only steps 2, 5 and 7 to obtain new
stable versions of aips++.