
 News News |
 FAQ FAQ
|
 Search Search
|
 Home Home
|
| Getting Started | Documentation | Glish | Learn More | Programming | Contact Us |

| Version 1.9 Build 1488 |
|
Tim Cornwell/Joe McMullin
This chapter describes how to make continuum OTF single dish images. It should be read in conjunction with several other chapters in Getting Results, including the chapters which cover Display of data and Image analysis.
The examples here are worked from GBT data but the principles apply to other telescopes. In particular, this chapter provides a facile means of reducing some standard mode continuum images. Different modes/analysis can still be obtained through the direct use of the basic AIPS++ tools.
GBT data are read into an AIPS++ MeasurementSet using the gbtmsfiller tool.
include 'gbtmsfiller.g'
filler := gbtmsfiller()
filler.setproject ('/home/gbtdata/pnt_prime_13')
filler.fillall()
filler.done()
This will create a MeasurementSet called pnt_prime_13_DCR in the current directory.
This can also be done from the command line before entering the AIPS++ shell.
> gbtmsfiller project=/home/gbtdata/zeroSpacing01 minscan=101 maxscan=301 msrootname='orion_' fillrawpointing=True
This will create a MeasurementSet called orion_DCR in the current directory (it appends the name of the backend, in this case the GBT DCR).
The continuum calibration imaging utilities are currently packaged under a single script which can be optionally added to an aips++ session. This script is called 'gbtcalutils.g'. It can be loaded upon startup as:
> aips++ -l gbtcalutils.g
For a minimal startup (only opens the necessary AIPS++ utilities required for continuum processing), one can type:
> glish -l gbtcalutils.g
Or it can be loaded to an existing session of AIPS++:
- include 'gbtcalutils.g'
This script makes a tool, gc (Gbt Continuum imaging), which has several functions appropriate for calibrating and imaging GBT continuum data; the tool assumes that you already have available a MeasurementSet (MS) of the observed map.
Interactive editing is possible on almost all graphical views of the data which are possible in msplot. To enable the editing option, it is necessary to set parameter edit=T when the msplot tool is created.
include 'msplot.g'
msp := msplot('pnt_prime_13_DCR', edit=T)
The GUI panel controlling flagging in msplot allows global flag masks to be set, such as that for all polarizations if any one is bad, as well as access to the list of accumulated flags, reversal of flags already applied, and other related utility operations. Raster displays of data are possible also, and are recommended for spectral-line data editing.
You may also view and edit the data at a lower level: that of the tables themselves. To browse the MeasurementSet table using a graphical browser, do:
include 'table.g'
t := table('pnt_prime_13_DCR')
t.browse()
Note that editing of individual values using the browser is possible (but unlikely to be needed unless there is something structurally wrong with the tables).
As noted above, AIPS++ data are stored in a MeasurementSet (MS). The observed data are stored in the DATA (complex) or FLOAT_DATA (real) column in the MAIN table of the MS; these are the raw data as loaded from the telescope archive or from a UVFITS file. Associated with the DATA column are related columns to hold the most recent version of the calibrated data (CORRECTED_DATA), and the latest data plane representation of the source or field model (MODEL_DATA). The latter two columns are populated at the filling stage (for GBT data these are initially just copies of the raw FLOAT_DATA column) or upon invocation of the imager tool on the MS. The observed data do not change during reduction, but the related columns do, and should be viewed in this context. When plotting the data using a tool such as msplot, the data sub-types (observed, corrected and model) can be selected individually.
An example data set checked into the AIPS++ system can be converted into a MS by using the following from the Unix prompt:
> gbtmsfiller project=/home/aips++/data/nrao/GBT/pnt_prime_13 minscan=2350 \
maxscan=2424 msrootname='cygnus_' fillrawpointing=True
This creates a MS, cygnus_DCR. Another data set which has already been converted into a data set is also available: /aips++/data/demo/dishdemo/pnt_lowgreg_11_DCR. Either of these can be used in the following examples.
setdata takes a single argument, the MeasurementSet name, and
examines it for suitability (e.g., is it a GBT continuum map) and content,
i.e., does it contain all of the requisite information to reduce automatically.
In particular, it explores the data set to confirm that information on the
polarization, bandwidth, and frequency are all present. In addition, it also
obtains the extent of the map and calculates the appropriate cell size for
gridding (hardwired to ![]() /2D).
/2D).
The initial continuum maps obtained at the GBT are four phase data representing the states of noise-diode off and on for two separate receptors. contcal performs the following calculation for each OTF strip of the map:
TANT = TCAL x 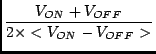 - < TCAL/2. >
- < TCAL/2. >
|
(5.1) |
V_ON are the counts for the CAL ON phase V_OFF are the counts for the CAL OFF phase T_CAL is the K/counts conversion
Imaging is done with the Imager tool, which supports a broad range of imaging modes and deconvolution algorithms, as described in the Imaging chapter of Getting Results and in the User Reference Manual.
The Imager tool requires initial selection of the data and initialization of the imaging coordinates.
A single-field, continuum GBT unnormalized image can be made using:
makeimage performs the steps to select, regrid and construct the image. It accepts arguments to specify the output image, the gridding function to be used and which receptor's data to image. In addition, an image of the coverage (sampling) is made which is simply the image name with a "_weight" appended to it. The coverage image is simply the gridded weights. The receptor is a numeric designation for the polarizations to which each receiver is sensitive (e.g., XX, YY, etc). If contcal was executed with average=T, then this will be an average of these polarizations; otherwise, each can be imaged separately.
The process of making a single dish image is very straightforward. The data are converted into the coordinate system of the image (set via Imager.setimage), and then added to the image using a gridding function. A variety of convolution functions can be used: BOX, SF, or PB. The first is a simple nearest neighbor gridding, the second uses a prolate spheroidal wavefunction, and the last uses a primary beam model appropriate to the telescope being used. The primary beam is optimal in the least squares sense but degrades the resolution, whereas the prolate spheroidal function can avoid the resolution degradation but the noise level will be higher.
The image from makeimage is not normalized by the sampling density. covercorr corrects for this by correcting for the coverage image to avoid nonsampled points (hardwired to the 10% level of the coverage map). If makeimage was previously executed, the values of "imname" (the source image) and "wname" (the weights image) are set. They may also be specified explicitly. The output image will be the name from "imname" with a "_corr" concatenated onto the end.
At this point, an image has been made which can be viewed simply by:
- im:=image('scanimage_corr')
- im.view();
plotsource also enables viewing of the contstructed images. It takes as an argument the name of the image to view and an optional parameter for the expected number of sources. The image module supports a findsources utility which identifies the positions of strong point sources in an image; it labels the position with 'Source x', where x is the number of the source found.
At this stage, use the viewer to manipulate the image in any way desired. The 'Adjust' button on the lower left provides the controls to the raster, contour and skycatalog information.
> cp -r /home/aips++/data/demo/dishdemo/pnt_lowgreg_11_DCR .
> glish -l gbtcalutils.g
NORMAL: gbtcalutils (gc) is ready for use
- field_names(gc) # look at the available tools in gc
# gc = GBT continuum imaging
checkms setdata contcal makeimage covercorr plotsource debug
- gc.setdata('cassaDCR');
successful readonly open of default-locked table cassaDCR/NRAO_GBT_GLISH: 21 col
umns, 120 rows
successful readonly open of default-locked table cassaDCR/NRAO_GBT_IF: 22 column
s, 240 rows
successful read/write open of default-locked table cassaDCR/FEED: 12 columns, 2
rows
successful read/write open of default-locked table cassaDCR/POLARIZATION: 4 colu
mns, 2 rows
successful read/write open of default-locked table cassaDCR/SPECTRAL_WINDOW: 14
columns, 1 rows
successful readonly open of default-locked table cassaDCR/POINTING: 15 columns,
48000 rows
T
- gc.contcal()
successful read/write open of default-locked table cassaDCR: 28 columns, 192000
rows
T
- gc.makeimage()
successful readonly open of default-locked table cassaDCR: 28 columns, 192000 ro
ws
Starting server imager
Server started: /export/home/guthwine/aips++/linux_gnu/bin/imager (AIPS++ versio
n: 1.7 (build #405))
Opening MeasurementSet cassaDCR
defaultcatalog (dc) ready
Starting imager::setdata
Selecting data
Performing selection on MeasurementSet
Selecting on field and spectral window ids
By selection 192000 rows are reduced to 48000
Finished imager::setdata
0.96 real 0.69 user 0.19 system
successful readonly open of default-locked table cassaDCR/NRAO_GBT_GLISH: 21 col
umns, 120 rows
Cell size is 0.289495253arcmin
Map size is 117.705376 by 59.9669179
Grid sizes are 406 208
Defining image properties
nx = 406 is not composite; nx = 432 or 400 will be more efficient
ny = 208 is not composite; ny = 216 or 200 will be more efficient
Setting processing options
Starting imager::weight
Weighting MS: IMAGING_WEIGHT column will be changed
Natural weighting
Sum of weights = 48000
Finished imager::weight
32.6 real 23.58 user 6.11 system
Starting imager::makeimage
Calculating image (without full skyequation)
Making single dish image from corrected data
Image is : scanimage
Frequency = 8.9 GHz, synthesized continuum bandwidth = 0.08 GHz
Image polarization = Stokes I
Frequency = 8.9 GHz, synthesized continuum bandwidth = 0.08 GHz
Image polarization = Stokes I
Performing Single Dish gridding with convolution function SF
Gridding will use specified common tangent point:
23:21:00.00 +58.32.00.00 B1950
Using prolate spheriodal wave function as the convolution function
Finished imager::makeimage
10.25 real 6.77 user 0.49 system
Starting imager::makeimage
Calculating image (without full skyequation)
Making single dish coverage function
Image is : scanimage_weight
Frequency = 8.9 GHz, synthesized continuum bandwidth = 0.08 GHz
Image polarization = Stokes I
Frequency = 8.9 GHz, synthesized continuum bandwidth = 0.08 GHz
Image polarization = Stokes I
Using prolate spheriodal wave function as the convolution function
Finished imager::makeimage
8.12 real 6.58 user 0.52 system
Successfully closed empty server: imager
T
- gc.covercorr()
Starting server app_image
Server started: /export/home/guthwine/aips++/linux_gnu/bin/app_image (AIPS++ ver
sion: 1.7 (build #405))
Selected bounding box :
[1, 1, 1, 1] to [406, 208, 1, 1] (23:28:23.897, +58.01.04.977, I, 8.900000e
+09Hz to 23:13:25.534, +59.00.59.643, I, 8.900000e+09Hz)
Creating new statistics storage lattice of shape [9]
Number points = 8.444800e+04 Sum = 8.444801e+04
Mean = 1.000000e+00
Variance = 1.026810e+00 Std dev = 1.013316e+00
Rms = 1.423657e+00
Minimum value 0.000000e+00 at [1, 1, 1, 1] (23:28:23.897, +58.01.04.977, I, 8.
900000e+09Hz)
Maximum value 4.499732e+00 at [105, 182, 1, 1] (23:24:41.961, +58.54.05.639, I
, 8.900000e+09Hz)
Creating image `scanimage_corr' of shape [406, 208, 1, 1]
Created mask `mask0'
T
- #Scale images to Jy (e.g., 1.76 K/Jy)
- im:=imagecalc(outfile='scanimage_tmb',pixels='(scanimage_corr)/1.76');
- im.view(); #note the higher amplitudes
- #You can also modify/add header information to the image
- #See the images module for more details; below is an example of adding
- #information on the restoring beam
- rb:=[=];
- rb.major.value:=87.6;
- rb.major.unit :='arcsec';
- rb.minor.value:=87.6;
- rb.minor.unit :='arcsec';
- rb.positionangle.value:=0;
- rb.positionangle.unit :='deg'
- im.setrestoringbeam(beam=rb);
- im.restoringbeam(); #see restoring beam values
- im.setbrightnessunit('Jy/beam');
- im.brightnessunit(); #see brightness unit value